生命理工学系 News
【研究室紹介】 田口研究室
シャペロン研究を軸にタンパク質フォールディングの謎に迫る
生命理工学系にはライフサイエンスとテクノロジーに関連した様々な研究室があり、基礎科学と工学分野の研究のみならず、医学や薬学、農学等、幅広い分野で最先端の研究が活発に展開されています。
研究室紹介シリーズでは、ひとつの研究室にスポットを当てて研究テーマや研究成果を紹介。今回は、プリオンやアミロイドなどタンパク質凝集体を研究する、田口研究室です。

主担当 生命理工学コース
教授 田口 英樹![]()
| キーワード | タンパク質、シャペロン、プリオン、アミロイド |
|---|---|
| Webサイト | 田口研究室 |
研究紹介
生命が生きていくためにはタンパク質が必要です。タンパク質がはたらくためには、アミノ酸がつながってできた「ひも」がある特定の「かたち」に折れたたむ(フォールディング)必要があります。フォールディングに失敗するとタンパク質は凝集体になってしまいますが、細胞の中では「シャペロン」というタンパク質がフォールディングを助けています。私たちは、シャペロンがどのような仕組みではたらいているのか、凝集体はどのようなメカニズムでできるのかなどを調べることで、フォールディングの秘密を解明したいと考えています。また、細胞内でタンパク質ができるプロセス(「翻訳」と呼ばれます)でどのようにフォールディングが起こるのか、など翻訳時の新生ポリペプチド鎖(新生鎖)の研究も精力的に進めています(新生鎖の生物学)。さらに、タンパク質の「かたち」が崩れることが病気に関係することがあります。代表例は狂牛病などでおなじみの「プリオン」です。パン酵母で見つかったプリオン的な因子をモデルにしたプリオンの研究も行っています。
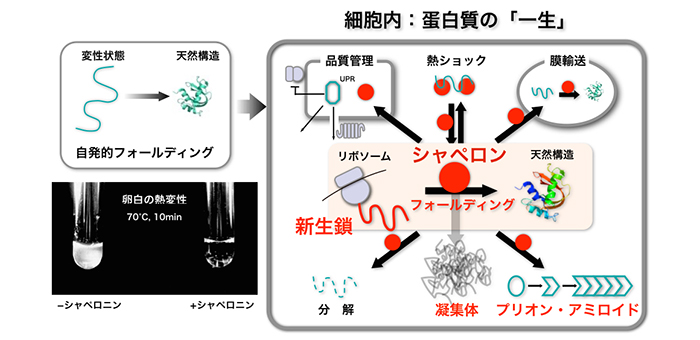
研究内容図
研究成果
代表論文
分子シャペロン、タンパク質の凝集体形成機構
- [1] Chadani Y, Niwa T, Chiba S, Taguchi, H.*, Ito K.* Integrated in vivo and in vitro nascent chain profiling reveals widespread translational pausing. Proc Natl Acad Sci USA. 113, E829-38 (2016)
- [2] Okuda, M, Niwa, T., *Taguchi, H.* Single-molecule analyses on the dynamics of Hsp104 and protein aggregates. J. Biol. Chem. 290, 7833-7840 (2015)
- [3] Ishimoto, T., Fujiwara, K., Niwa, T., Taguchi, H.*: Conversion of a chaperonin GroEL-independent protein into an obligate substrate. J. Biol. Chem. 289 32073-32080 (2014)
- [4] Taguchi, H.* Reaction cycle of chaperonin GroEL via symmetric "football" intermediate (review). J. Mol. Biol. 427, 2912-2918 (2015)
- [5] Koike-Takeshita, A., Mitsuoka, K., Taguchi, H.* Asp52 in combination with Asp398 plays a critical role in ATP hydrolysis of chaperonin GroEL. J. Biol. Chem. 289 30005-30011 (2014)
- [6] Niwa, T., Kanamori T., *Ueda, T., *Taguchi, H. Global Analysis of Chaperone Effects Using a Reconstituted Cell-Free Translation System. Proc. Natl. Acad. Sci. U.S.A. 109, 8937-8942 (2012)
- [7] Fujiwara, K., Ishihama, Y., Nakahigashi, K., Soga, T. and Taguchi, H.* A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 29, 1552-1564 (2010)
- [8] Niwa, T., Ying, B.-W., Saito, K., Jin, W. Z., Takada, S., Ueda, T. Taguchi, H.* Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 4201-4206 (2009)
- [9] #Ueno, T., #Taguchi, H., Tadakuma, H., Yoshida, M., Funatsu, T. GroEL mediates protein folding with a two successive timer mechanism. Mol. Cell 14, 423-434 (2004)
- [10] Taguchi, H., Ueno, T., Tadakuma, H., Yoshida, M., Funatsu, T. Single-molecule observation of protein-protein interactions in the chaperonin system. Nat. Biotechnol. 19, 861-865 (2001)
酵母プリオン
- [1] Odani, W, Urata, K, Okuda, M, Okuma, S, Koyama, H, Pack, CG, Fujiwara, K, Nojima, T, Kinjo, M, Kawai-Noma, S, Taguchi, H.* Peptide sequences converting polyglutamine into a prion in yeast. FEBS J. 282, 477-490 (2015)
- [2] Kawai-Noma, S., Pack, C-G., Kojidani, T., Asakawa, H., Hiraoka, Y., Kinjo, M., Haraguchi, T., Taguchi, H.*, and Hirata, A. In vivo evidence for the fibrillar structures of Sup35 prions in yeast cells. J. Cell Biol. 190, 223-231 (2010)
- [3] Taguchi, H.* and Kawai-Noma, S. Diffuse oligomer-based transmission of yeast prions. (Review) FEBS J. 277, 1359-1368 (2010)
- [4] Kawai-Noma, S., Ayano, S., Pack, C-G., Kinjo, M., Yoshida, M., Yasuda, K., Taguchi, H. Dynamics of yeast prion aggregates in single living cells. Genes to Cells 11, 1085-1096 (2006)
- [5] Inoue, Y., Taguchi, H., Kishimoto, A., Yoshida, M. Hsp104 binds to yeast sup35 prion fiber but needs other factor(s) to sever it. J. Biol. Chem. 279, 52319-52323 (2004)
- [6] Inoue, Y., Kishimoto, A. et al. Strong growth polarity of yeast prion-fiber revealed by single fiber imaging. J. Biol. Chem. 276, 35227-35230 (2001)
主な日本語総説
- [1] 田口 英樹「タンパク質フォールディングの「理想」と「現実」:凝集形成とシャペロンの役割」生化学 87, 194-204 (2015)
- [2] 丹羽達也、上田卓也、田口 英樹「大腸菌全タンパク質の凝集性とシャペロン効果の網羅的な解析」生物物理 53, 309-312 (2013)
- [3] 田口 英樹「タンパク質のフォールディング:理想と現実」高分子 62, 502-504 (2013)
著書
- [1] 「池上彰が聞いてわかった生命のしくみ」池上彰、岩崎博史、田口英樹著、朝日新聞出版、2016年
- [2] 「学んでみると生命科学はおもしろい」田口英樹著、ベレ出版2014年
教員紹介
田口英樹 教授 博士(理学)
| 1993年9月 | 東京工業大学 大学院総合理工学研究科 生命化学専攻 博士課程修了 |
|---|---|
| 1993 - 1995年 | 日本学術振興会 特別研究員 |
| 1995 - 2003年 | 東京工業大学 資源化学研究所 助手 |
| 2003 - 2010年 | 東京大学 大学院新領域創成科学研究科 准教授 |
| 2010 - 2016年 | 東京工業大学 大学院生命理工学研究科 教授 |
| 2016年4月より | 科学技術創成研究院 細胞制御工学研究ユニット(生命理工学院担当)教授 |
| 2002年11月より | 科学技術振興機構さきがけ研究者を兼任 |
| 2013年 | 手島記念研究賞(論文賞) |
|---|
- 教育活動
学部:生命科学基礎第二、生物物理化学
大学院:細胞物理生物学
- 所属学会
- 日本蛋白質科学会(理事)、日本生化学会、日本生物物理学会、日本分子生物学会、日本細胞生物学会
教員からのメッセージ
- 田口教授より
-
Let's Enjoy Science!をモットーに、今まで世界の誰も知らないことを、新しいアイディア、新しい研究手法、新しい材料で研究していきたいと考えています。拡張し続けているタンパク質の世界にどっぷり浸かって、自ら考えてアイディアを出したいと思っている人は一緒にサイエンスを楽しみましょう。「楽しむ」というのは「楽(らく)」というのとは違いますが、基礎研究に没頭できる環境を準備してお待ちしています。
- 研究室と研究テーマ
- タンパク質合成過程における「緩急のリズム」を実証―大腸菌遺伝子産物の中間状態を網羅的に解析―|東工大ニュース
- 顔 東工大の研究者たち Vol.10 田口英樹|研究ストーリー|研究
- 科学技術創成研究院 細胞制御工学研究ユニット
お問い合わせ先
教授 田口英樹
すずかけ台キャンパス S2棟 6階
E-mail : taguchi@bio.titech.ac.jp
※この内容は掲載日時点の情報です。最新の研究内容については研究室サイト![]() をご覧ください。
をご覧ください。
※ 2025年5月1日:一部最新の情報に更新しました。