Life Science and Technology News
【Labs spotlight】 Taguchi Laboratory
Pursuing the mechanism of chaperone and prion
The Department has a variety of laboratories for Life Science and Technology, in which cutting-edge innovative research is being undertaken not only in basic science and engineering but also in the areas of medicine, pharmacy, agriculture, and multidisciplinary sciences.
This "Spotlight" series features a laboratory from the Department and introduces you to the laboratory's research projects and outcomes. This time we focus on Taguchi Laboratory.

Areas of Supervision
Primary/Life Science and Technology
Professor Hideki Taguchi![]()
| Degree | PhD 1993, Tokyo Institute of Technology |
|---|---|
| Areas of Research | Biochemistry, Biophysics, Protein Sciences. |
| Keywords | Chaperones, Protein Folding, Protein Aggregation, Prion/Amyloid, Nascent chain biology. |
| Website | Taguchi Laboratory |
Research interest
Life depends on proteins. Proteins are linear chains of amino acids (polypeptide chains) that adopt unique three-dimensional structures. The polypeptide chains, which are emerged from the ribosome in the cell, must fold into the tertiary structures for biological functions. Although the folding process is spontaneous, protein folding is often hampered by inter-molecular protein aggregation, which can be prevented by a variety of chaperone proteins in the cell. Our lab interests are focused on the molecular mechanisms that affect the protein folding in the cell. How chaperones assist in the protein folding in the cell? Which proteins are aggregation-prone? What are the determinants that define the aggregation-prone propensity and the chaperone dependency?
In addition, we are also working on the biology of prion/amyloids. A subset of proteins forms amyloids, which are ordered aggregates composed of cross-beta sheet structures. Amyloid aggregates are associated with various human neurodegenerative diseases, including Alzheimer's disease and Huntington's disease. Amyloids include prions, defined as infectious proteins, which are responsible for mammalian transmissible spongiform encephalopathies such as mad-cow disease. The concept of prions has been expanded to include several epigenetically heritable traits in fungi, the yeast prions. We are pursuing to elucidate the mechanism underlying the propagation of prions using yeast prions as a model.
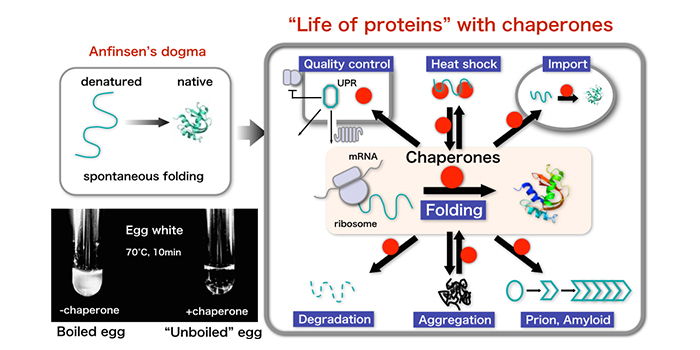
Research findings
Selected publications
Chaperones, Protein aggregation
- [1] Chadani Y, Niwa T, Chiba S, Taguchi, H.*, Ito K.* Integrated in vivo and in vitro nascent chain profiling reveals widespread translational pausing. Proc Natl Acad Sci USA. 113, E829-38 (2016)
- [2] Okuda, M, Niwa, T., *Taguchi, H.* Single-molecule analyses on the dynamics of Hsp104 and protein aggregates. J. Biol. Chem. 290, 7833-7840 (2015)
- [3] Ishimoto, T., Fujiwara, K., Niwa, T., Taguchi, H.*: Conversion of a chaperonin GroEL-independent protein into an obligate substrate. J. Biol. Chem. 289 32073-32080 (2014)
- [4] Taguchi, H.* Reaction cycle of chaperonin GroEL via symmetric "football" intermediate (review). J. Mol. Biol. 427, 2912-2918 (2015)
- [5] Koike-Takeshita, A., Mitsuoka, K., Taguchi, H.* Asp52 in combination with Asp398 plays a critical role in ATP hydrolysis of chaperonin GroEL. J. Biol. Chem. 289 30005-30011 (2014)
- [6] Niwa, T., Kanamori T., *Ueda, T., *Taguchi, H. Global Analysis of Chaperone Effects Using a Reconstituted Cell-Free Translation System. Proc. Natl. Acad. Sci. U.S.A. 109, 8937-8942 (2012)
- [7] Fujiwara, K., Ishihama, Y., Nakahigashi, K., Soga, T. and Taguchi, H.* A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 29, 1552-1564 (2010)
- [8] Niwa, T., Ying, B.-W., Saito, K., Jin, W. Z., Takada, S., Ueda, T. Taguchi, H.* Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 4201-4206 (2009)
- [9] #Ueno, T., #Taguchi, H., Tadakuma, H., Yoshida, M., Funatsu, T. GroEL mediates protein folding with a two successive timer mechanism. Mol. Cell 14, 423-434 (2004)
- [10] Taguchi, H., Ueno, T., Tadakuma, H., Yoshida, M., Funatsu, T. Single-molecule observation of protein-protein interactions in the chaperonin system. Nat. Biotechnol. 19, 861-865 (2001)
Yeast prions
- [1] Odani, W, Urata, K, Okuda, M, Okuma, S, Koyama, H, Pack, CG, Fujiwara, K, Nojima, T, Kinjo, M, Kawai-Noma, S, Taguchi, H.* Peptide sequences converting polyglutamine into a prion in yeast. FEBS J. 282, 477-490 (2015)
- [2] Kawai-Noma, S., Pack, C-G., Kojidani, T., Asakawa, H., Hiraoka, Y., Kinjo, M., Haraguchi, T., Taguchi, H.*, and Hirata, A. In vivo evidence for the fibrillar structures of Sup35 prions in yeast cells. J. Cell Biol. 190, 223-231 (2010)
- [3] Taguchi, H.* and Kawai-Noma, S. Diffuse oligomer-based transmission of yeast prions. (Review) FEBS J. 277, 1359-1368 (2010)
- [4] Kawai-Noma, S., Ayano, S., Pack, C-G., Kinjo, M., Yoshida, M., Yasuda, K., Taguchi, H. Dynamics of yeast prion aggregates in single living cells. Genes to Cells 11, 1085-1096 (2006)
- [5] Inoue, Y., Taguchi, H., Kishimoto, A., Yoshida, M. Hsp104 binds to yeast sup35 prion fiber but needs other factor(s) to sever it. J. Biol. Chem. 279, 52319-52323 (2004)
- [6] Inoue, Y., Kishimoto, A. et al. Strong growth polarity of yeast prion-fiber revealed by single fiber imaging. J. Biol. Chem. 276, 35227-35230 (2001)
- Research Laboratories and Subjects
- New insights into Protein Synthesis | Tokyo Tech News
- FACES: Tokyo Tech Researchers, Issue 10 - Hideki Taguchi | Research Stories | Research
- Cell Biology Unit
Contact
Professor TaguchiTaguchi
Floor 6, S2 building, Suzukakedai campus
E-mail : taguchi@bio.titech.ac.jp
*Find more about the lab and the latest activities at the lab site![]() .
.
*May 1, 2025:Some of the content has been updated with the latest information.