Life Science and Technology News
Engineering Bacteria to Biosynthesize Intricate Protein Complexes
Protein cages found in nature within microbes help weather its contents from the harsh intracellular environment—an observation with many bioengineering applications. Tokyo Tech researchers recently developed an innovative bioengineering approach using genetically modified bacteria; these bacteria can incorporate protein cages around protein crystals. This in-cell biosynthesis method efficiently produces highly customized protein complexes, which could find applications as advanced solid catalysts and functionalized nanomaterials.
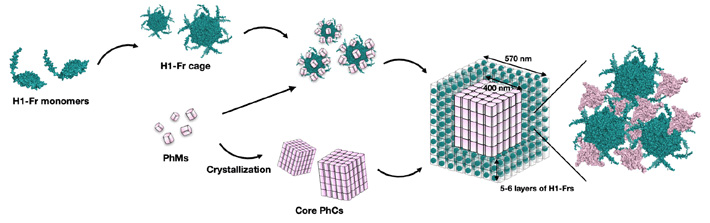
Figure 1. In-cell assembly process of H1-Fr/PhC
- This diagram shows how H1-Fr monomers and polyhedrin monomers (PhMs) combine to spontaneously form a complex core–shell structure inside the E. coli bacteria
In nature, proteins can assemble to form organized complexes with myriad shapes and purposes. Thanks to the remarkable progress in bioengineering over the past few decades, scientists can now produce customized protein assemblies for specialized applications. For example, protein cages can confine enzymes that act as catalysts for a target chemical reaction, weathering it from a potentially harsh cell environment. Similarly, protein crystals—structures composed of repeating units of proteins—can serve as scaffolds for synthesizing solid materials with exposed functional terminals.
However, incorporating (or 'encapsulating') foreign proteins on the surface of a protein crystal is challenging. Thus, synthesizing protein crystals encapsulating foreign protein assemblies has been elusive. So far, no efficient methods exist to achieve this goal, and the types of protein crystals produced are limited. But what if bacterial cellular machinery can achieve this goal?
In a recent study, a research team from Tokyo Institute of Technology, including Professor Takafumi Ueno, reported a new in-cell method for encapsulating protein cages with diverse functions on protein crystals. Their paper, published in Nano Letters![]() , represents a substantial breakthrough in protein crystal engineering.
, represents a substantial breakthrough in protein crystal engineering.
The team's innovative strategy involves genetically modifying Escherichia coli bacteria to produce two main building blocks: polyhedrin monomer (PhM) and modified ferritin (Fr). On the one hand, PhMs naturally combine within cells to form a well-studied protein crystal called polyhedra crystal (PhC). On the other hand, 24 Fr units are known to combine to form a stable protein cage. "Ferritin has been widely used as a template for constructing bio-nano materials by modifying its internal and external surfaces. Thus, if the formation of a Fr cage and its subsequent immobilization onto PhC can be performed simultaneously in a single cell, the applications of in-cell protein crystals as bio-hybrid materials will be expanded," explains Prof. Ueno.
To immobilize the Fr cages into PhC, the researchers modified the gene coding for Fr to include an α-helix(H1) tag of PhM, thus creating H1-Fr. The reasoning behind this approach is that the H1-helixes naturally present in PhM molecules interact significantly with the tags on H1-Fr, acting as 'recruiting agents' that bind the foreign proteins onto the crystal.
Using advanced microscopy, analytical, and chemical techniques, the research team verified the validity of their proposed approach. Through various experiments, they found that the resulting crystals had a core–shell structure, namely a cubic PhC core about 400 nanometers wide covered in five or six layers of H1-Fr cages.
This strategy for the biosynthesis of functional protein crystals holds much promise for applications in medicine, catalysis, and biomaterials engineering. "H1-Fr cages have the potential to immobilize external molecules inside them for molecular delivery," remarks Prof. Ueno, "Our results indicate that the H1-Fr/PhC core–shell structures, displaying H1-Fr cages on the outer surface of the PhC core, can be individually controlled at the nanoscale level. By accumulating different functional molecules in the PhC core and H1-Fr cage, hierarchical nanoscale-controlled crystals can be constructed for advanced biotechnological applications."
Future works in this field will help us realize the true potential of bioengineering protein crystals and assemblies. With any luck, these efforts will pave the way to a healthier and more sustainable future.
- Reference
| Authors : | Thuc Toan Pham1, Satoshi Abe1, Koki Date1, Kunio Hirata2, Taiga Suzuki1, and Takafumi Ueno1,3 |
|---|---|
| Title : | Displaying a Protein Cage on a Protein Crystal by In-Cell Crystal Engineering | Journal : | Nano Letters |
| DOI : | 10.1021/acs.nanolett.3c02117 |
| Affiliations : |
1 School of Life Science and Technology, Tokyo Institute of Technology 2 SR Life Science Instrumentation Unit, RIKEN/SPring-8 Center 3 Living Systems Materialogy (LiSM) Research Group, International Research Frontiers Initiative (IRFI), Tokyo Institute of Technology |
- Towards Artificial Photosynthesis with Engineering of Protein Crystals in Bacteria | Life Science and Technology News
- Novel Cell-Free Protein Crystallization Method to Advance Structural Biology | Life Science and Technology News
- Tokyo Tech Research Video "Dynamic self-assembly of designed protein 'Needles'" Now Online | Life Science and Technology News
- Cats in a Cage: Novel Hybrid Nanocages for Faster Catalysis | Life Science and Technology News
- Decoding Protein Assembly Dynamics with Artificial Protein Needles | Life Science and Technology News
- In-cell Nano-3D Printer: Synthesizing Stable Filaments from In-cell Protein Crystals | Life Science and Technology News
- Joint Research Coronavirus Task Force Established | Life Science and Technology News
- Nanotubes built from protein crystals: Breakthrough in biomolecular engineering | Life Science and Technology News
- Nanocages for gold particles: what is happening inside? | Life Science and Technology News
- In cell molecular sieve from protein crystal | Life Science and Technology News
- Ferritin releases carbon monoxide in regulated therapeutic doses | Tokyo Tech News
- Protein-engineered cages aid studies of cell functions | Tokyo Tech News
- Dynamic self-assembly of designed protein 'Needles' | YouTube
- Satoshi Abe | Researcher Finder - Tokyo Tech STAR Search
- Takafumi Ueno | Researcher Finder - Tokyo Tech STAR Search
- Ueno Laboratory
- Department of Life Science and Technology, School of Life Science and Technology
- Latest Research News
School of Life Science and Technology
—Unravel the Complex and Diverse Phenomena of Life—
Information on School of Life Science and Technology inaugurated in April 2016
Further Information
Assistant Professor Satoshi Abe
School of Life Science and Technology,
Tokyo Institute of Technology
Email saabe@bio.titech.ac.jp
Tel +81-45-924-5806
Professor Takafumi Ueno
School of Life Science and Technology,
Tokyo Institute of Technology
Email tueno@bio.titech.ac.jp
Tel +81-45-924-5844