Life Science and Technology News
Untapped Power: Logical Operations Using RNA Droplets
Researchers make strides in the use of RNA as a biocomputational tool
RNA droplets can now be used to perform logical operations that take microRNA sequences as inputs, report scientists from Tokyo Tech. By self-assembling into network-like structures, RNA molecules form liquid-state droplets. These RNA droplets disperse only when the correct microRNA sequences are present by performing the logical AND operation. This innovative strategy could pave the way to advances in biomolecular sensing, artificial cells, and computational biodevices.
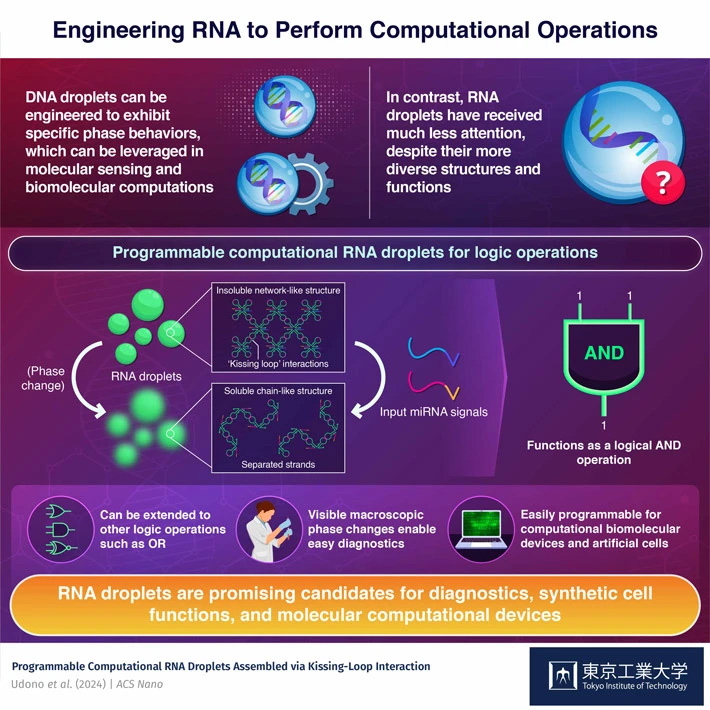
Researchers introduce computational RNA droplets capable of performing two-input AND logic operations, offering a flexible approach for programmable assembly of biomolecular devices and artificial cells.
DNA and RNA encode genetic information and are involved in protein synthesis. In DNA/RNA nanotechnology, their information-processing capabilities are harnessed to form programmable 2D/3D structures. In recent years, DNA droplets, liquid-state condensates of synthetic DNA, have attracted increasing attention as molecular sensing and biocomputational devices. While DNA droplets have captured the spotlight in this advanced research field, their RNA counterparts can be just as powerful, if not more. Compared to DNA, RNA has much more varied functions besides its role in protein synthesis. Additionally, it can adopt a broad array of molecular structures. Unfortunately, despite its versatility, few studies have explored the untapped potential of RNA droplets as a tool for biocomputing and bio-nanotechnology.
Against this backdrop, a team of researchers, including Professor Masahiro Takinoue and post-doctoral researcher Hirotake Udono from Tokyo Institute of Technology, Japan, set out to develop a new type of 'computational RNA droplets,' which can be used to perform logic operations akin to electronic devices. Their study findings![]() were published on June 3 in ACS Nano and selected as supplementary cover.
were published on June 3 in ACS Nano and selected as supplementary cover.
These programmable RNA droplets are constructed through RNA's capabilities for sequence recognition and secondary structure formation. "Our approach is based on the kissing loop (KL) interaction, which occurs between two internally folded single-stranded RNAs," explains Takinoue. "This enables the self-assembly of these folded structures into stable complex structures." Simply put, the researchers carefully engineered the nucleotide sequence of the RNA strands so that they would spontaneously assemble into predetermined molecular structures, which also attract each other to form a large-scale network.
The phase-state controllability of RNA droplets is underlain by a type of key-and-lock mechanism that can only be 'opened' by a specific microRNA sequence. Due to the way the RNA structures were designed, only when two given microRNA sequences are present at the same time will the network disassemble into chain-like structures. When this happens, it causes a noticeable change in the phase of the RNA droplets, which become more dispersed in the solution.
In computational terms, the proposed RNA droplets can effectively perform the logical AND operation, taking two microRNA sequences as inputs. The researchers also stated that designing similar RNA droplets for other logical operations is also possible. This versatility could prove useful in several applications, as Takinoue remarks: "Our computational RNA droplets can be applied to the in situ programmable assembly of computational biomolecular devices and artificial cells from transcriptionally derived RNA within biological or artificial cells."
On top of this, these RNA droplets hold much promise as convenient diagnostic tools. Since they can detect the presence of specific RNA sequences, they can be used to look for disease biomarkers. "Unlike previously reported submicroscopic RNA-based logic operators, the macroscopic phase change of our RNA droplets provides a readout for molecular sensing that can be distinguished with the naked eye," highlights Takinoue.
Overall, this study highlights RNA's dual role as both a tool and a building block for versatile bioengineering devices. Stay tuned for more breakthroughs in this rapidly evolving field!
- Reference
| Authors : | Hirotake Udono1*, Minzhi Fan1, Yoko Saito1, Hirohisa Ohno2, Shin-ichiro M. Nomura3, Yoshihiro Shimizu4, Hirohide Saito2, and Masahiro Takinoue1,5,6* |
|---|---|
| Title : | Programmable Computational RNA Droplets Assembled via Kissing-Loop Interaction |
| Journal : | ACS Nano |
| DOI : | 10.1021/acsnano.3c12161 |
| Affiliations : | 1 Department of Computer Science, Tokyo Institute of Technology, Japan 2 Department of Life Science Frontiers, Center for iPS Cell Research and Application, Kyoto University, Japan 3 Department of Robotics, Graduate School of Engineering, Tohoku University, Japan 4 Laboratory for Cell-Free Protein Synthesis, Center for Biosystems Dynamics Research, RIKEN, Japan 5 Department of Life Science and Technology, Tokyo Institute of Technology, Japan 6 Research Center for Autonomous Systems Materialogy (ASMat), Institute of Innovative Research, Tokyo Institute of Technology, Japan |
| * Corresponding authors' emails: | udono.h.aa@m.titech.ac.jp (H.U.), takinoue@c.titech.ac.jp (M.T.) |
- Novel Method for Early Disease Detection Using DNA Droplets|Life Science and Technology News
- The changing patterns of DNA microcapsules|Life Science and Technology News
- Quantitative Approach on Understanding How Epigenetic Switches Control Gene Expression|Life Science and Technology News
- Designing DNA From Scratch: Engineering the Functions of Micrometer-Sized DNA Droplets|Life Science and Technology News
- Giving Movement to The Micro: Novel Technique Moves Micro-particles using Electric Fields|Life Science and Technology News
- Scientists develop DNA microcapsules with built-in ion channels|Life Science and Technology News
- Formation of Artificial Cells with a Skeletal Support Reinforcement to withstand Application Realized | Tokyo Tech News
- Successful use of DNA as a computer in artificial cells - Realizing molecular robots that work in vivo - | Tokyo Tech News
- Creating artificial cells and molecular robots using DNA - Exploring the boundary between matter and life | Research Stories | Research
- Masahiro Takinoue | Researcher Finder - Tokyo Tech STAR Search
- Takinoue Laboratory
- Department of Computer Science, School of Computing
- Research Center for Autonomous Systems Materialogy (ASMat)
- Institute of Innovative Research (IIR)
- Center for iPS Cell Research and Application, Kyoto University
- School of Engineering, Tohoku University
- RIKEN
- Latest Research News
School of Computing —Creating the Future Information Society—
Information on School of Computing inaugurated in April 2016
Further Information
Professor Masahiro Takinoue
Department of Computer Science, Tokyo Institute of Technology
E-mail takinoue@c.titech.ac.jp