Life Science and Technology News
Advances in Understanding the Evolution of Stomach Loss in Agastric Fishes
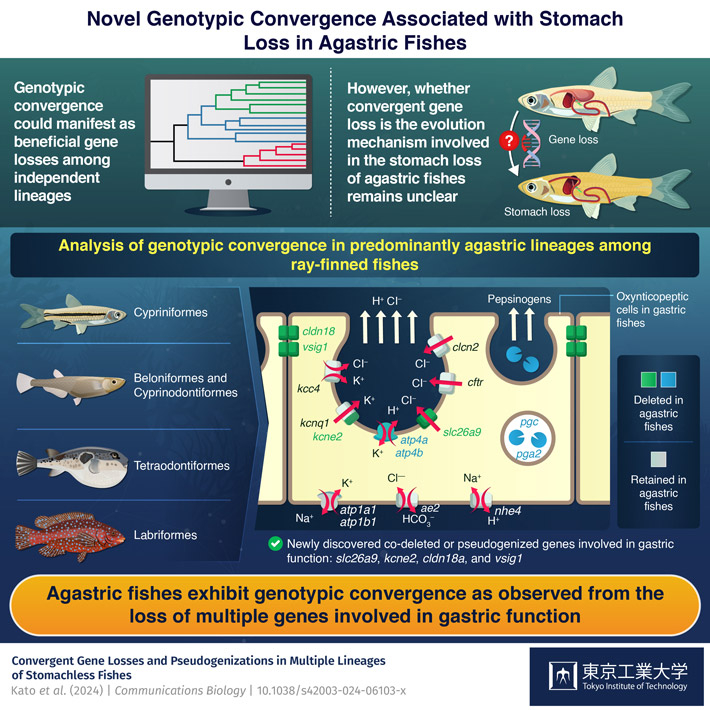
Genotypic convergence involving the loss of multiple genes required for gastric function could explain the evolution of different stomachless (or agastric) ray-finned fishes, according to a new study by Tokyo Tech researchers. Based on preliminary research identifying slc26a9 gene loss in agastric fishes compared to gastric fishes, the researchers further compared gene losses between agastric and gastric fishes to arrive at their findings, which revealed convergent gene losses and pseudogene-forming inactivations in agastric fishes.
Living beings can evolve to lose biological structures due to potential survival benefits from such losses. For example, certain groups of ray-finned fishes show such regressive evolution—medakas, minnows, pufferfishes, and wrasses do not have a stomach in the gastrointestinal tract, making them agastric or stomachless fishes. However, the specific evolutionary mechanisms underlying the evolution of agastric fishes remains unclear.
Studies about Slc26a9—a molecular transporter highly expressed in the stomach of many species—in fishes provided the initial clue. Researchers in Tokyo Institute of Technology (Tokyo Tech), Mayo Clinic College of Medicine, and Atmosphere and Ocean Research Institute, The University of Tokyo found that the slc26a9 gene was absent in many agastric ray-finned fishes but present in many gastric ray-finned fishes. These findings led them to ponder if more genes required for gastric function were absent in agastric fishes. Could such convergent gene losses account for stomach loss in agastric fishes?
A team of scientists from Japan and the USA, led by Associate Professor Akira Kato from Tokyo Tech, sought to answer this question. Kato explains, "We compared gene losses between agastric and gastric ray-finned fishes and identified additional genes co-deleted in agastric fishes." Accordingly, the researchers identified several genes required for gastric functions that are co-deleted or pseudogenized (gene inactivation through mutations resulting in pseudogenes) in agastric fishes compared to gastric fishes, namely slc26a9, kcne2, cldn18a, and vsig1. Their findings are published in Communications Biology![]() .
.
Specifically, they identified four gene deletions—slc26a9, kcne2, cldn18a, and vsig1—resulting in a reduction or loss of stomach structure in ray-finned fishes through comparative genomic analyses (a set of experiments for comparing similarities and differences among different genomes). Not surprisingly, each of these four genes codes for essential gastric functions. slc26a9 codes for the chlorine ion channel transporter. kcne2 codes for a regulatory subunit of the potassium ion channel. slc26a9 and kcne2 functions are, thus, essential for secretions of gastric acids, such as hydrochloric acid. cldn18a similarly codes for a barrier protecting the gastric cells from acid-induced damage from hydrogen ions, while vsig1 codes for controlling stomach development.
In addition, the researchers found that agastric egg-laying mammals, such as echidna and platypus, also have kcne2 and vsig1 either deleted or pseudogenized. Moreover, the researchers discovered that cldn18, if present in agastric bony fishes, is mutated compared to gastric fishes. All these findings indicate that the gene losses correlated with stomach loss represent a genotypic convergence.
Furthermore, they observed that the gastric fish, stickleback, expressed kcne2, pga, pgc, vsig1, and cldn18a in organs other than the stomach, indicating gene functions other than gastric functions. They deduced that agastric fishes could possess other genes to compensate for such gene functions, facilitating the observed gene losses.
Kato concludes, "We identified novel genes absent in agastric fishes among four major bony fish lineages, which suggests a convergent evolution scenario in the context of stomach loss. Our findings, thus, imply that a similar cassette of gene losses occurred independently during or after stomach loss in the several agastric fish groups." Indeed, this study is a milestone in understanding the novel genotypic convergence that fine-tuned the agastric fish body to suit its specific ecological niche.
- Reference
| Authors : | Akira Kato1,2,3,4*, Supriya Pipil5, Chihiro Ota1, Makoto Kusakabe5,6, Taro Watanabe5, Ayumi Nagashima1, An-Ping Chen4, Zinia Islam2, Naoko Hayashi2, Marty Kwok-Shing Wong5,7, Masayuki Komada1,8, Michael F. Romero4,9, and Yoshio Takei5 |
|---|---|
| Title : | Convergent gene losses and pseudogenizations in multiple lineages of stomachless fishes |
| Journal : | Communications Biology |
| DOI : | 10.1038/s42003-024-06103-x |
| Affiliations : |
1School of Life Science and Technology, Tokyo Institute of Technology, Japan 2Department of Biological Sciences (then), Tokyo Institute of Technology, Japan 3Center for Biological Resources and Informatics, Tokyo Institute of Technology, Japan 4Department of Physiology and Biomedical Engineering, Mayo Clinic College of Medicine and Science, USA 5Department of Marine Bioscience, Atmosphere and Ocean Research Institute, The University of Tokyo, Japan 6Department of Biological Sciences, Shizuoka University, Japan 7Department of Biomolecular Science, Toho University, Japan 8Cell Biology Center, Tokyo Institute of Technology, Japan 9Department of Nephrology and Hypertension, Mayo Clinic College of Medicine and Science, USA |
| * Corresponding author's email: | akirkato@bio.titech.ac.jp |
- New Study Sheds Light on Boric Acid Transport and Excretion in Marine Fish | Life Science and Technology News
- Absorbing sodium from fresh water in exchange with waste | Tokyo Tech News
- Akira Kato | Researcher Finder - Tokyo Tech STAR Search
- Kato Laboratory (Japanese)
- 【Labs spotlight】Kato Laboratory | Life Science and Technology News
- Department of Life Science and Technology, School of Life Science and Technology
- Mayo Clinic College of Medicine and Science
- Atmosphere and Ocean Research Institute, The University of Tokyo
- Latest Research News
School of Life Science and Technology
—Unravel the Complex and Diverse Phenomena of Life—
Information on School of Life Science and Technology inaugurated in April 2016
Further Information
Associate Professor Akira Kato
School of Life Science and Technology, Tokyo Institute of Technology