Life Science and Technology News
New study reveals life’s earliest evolution was more complicated than previously suspected
Biologists classify all living organisms into three major groups they call ‘domains’. Two of these domains—the Bacteria and the Archaebacteria—consist of single-celled organisms, while the third—the Eukaryota—includes most of the larger, multicellular organisms we are all familiar with: fungi, plants and animals including ourselves. Of the three domains, the Eukaryota almost certainly evolved the most recently, but questions remain about which of the two single-celled domains arose first in the history of life. Over forty years ago, American biologists Carl Woese and George Fox suggested that these two domains both emerged from a more primitive organism or group of organisms scientists now call LUCA, or the Last Universal Common Ancestor. Scientists would love to be able to say something concrete about what LUCA was like, what types of environment it lived in, and how it made its living. For example was it a heterotroph—an organism which eats environmentally supplied organic food—like ourselves, or an autotroph—an organism which is able to make its own food from inorganic compounds—like a photosynthesiser or the ‘rock-eating’ microbes that scientists call lithoautotrophs? If scientists could nail down some of these questions about LUCA, which they potentially could do by knowing which genes LUCA possessed, this might provide even deeper insights as to how and where life originated on Earth in the first place. New research from Tokyo Tech and the Max Planck Institute suggests understanding early life may be trickier than previously thought.
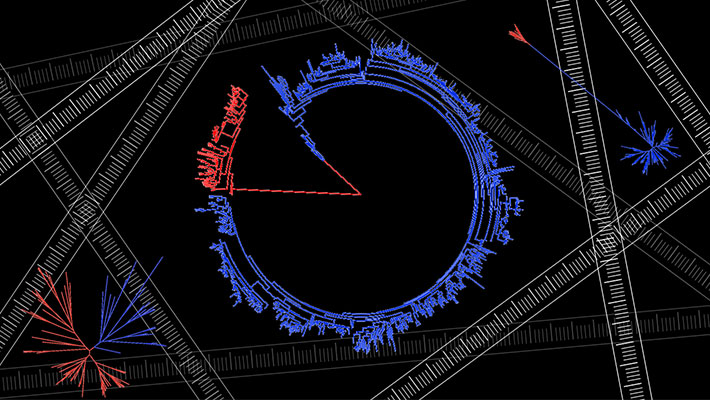
- Figure. 1
- Phylogenetic tree diagrams form the basis of understanding microbial evolution. Long branches between the two domains in some trees may reflect a period of very rapid evolution, billions of years ago.
Credit: S. Shiobara, S. McGlynn, ELSI, CC BY 4.0
Scientists can now easily sequence and compare the DNA of microorganisms, which allows them to determine how closely related organisms are. Just as we have our own family trees, organismal DNA comparisons are likewise a measure of which organisms shared common ancestors. Organisms with very similar DNA likely have recent common ancestors; the DNA from monkeys is similar to that from humans, and both monkeys and humans have DNA that is more similar to that of each other than that from octopuses, and so on. Those with less similar DNA sequences are likely more distantly related, and thus must have differentiated earlier. This sort of comparison allows scientists to reconstruct evolutionary relationships among organisms, and construct ‘family trees’ which relate them in time. DNA sequence comparison has revealed that all organisms on Earth are related to one another, and this allows scientists to build a ‘tree of life’, which shows how they are related evolutionarily.
There is, however, a major complication to this. Besides the normal parent-to-offspring inheritance of DNA which dominates in higher organisms such as ourselves, in microorganisms snippets of DNA can also ‘jump’ between organisms. This can occur by various means; for example, sometimes viral infections can move genes from one organism to another. It has been hypothesised that the earliest organisms may have undergone more rapid evolutionary change, partly because they were not able to copy information as accurately as modern ones can, but also due to more widespread ‘gene jumping’. If most ancient cells could not copy information as accurately as modern ones, and if their DNA moved more easily between lineages than today, using modern DNA to understand ancient evolution may be extremely misleading.
This is where new research carried out by Sarah Berkemer, based at the Max Planck Institute for Mathematics in the Sciences in Leipzig, Germany, and Shawn McGlynn from the Earth-Life Science Institute at the Tokyo Institute of Technology in Japan comes in. Their analyses confirm other work which suggested that only a limited understanding of the lifestyle of the most ancient cells can be derived from DNA comparison. Although this is a disappointing result for evolutionary biologists, it is important to understand what can and cannot be known from the data that scientists are able to gather from modern organisms. Berkemer and McGlynn’s work does supply one silver lining however; while it is clear that we don’t know what the first organisms metabolised or where they lived, their work provides insight into how quickly they may have evolved billions of years ago.
Berkemer and McGlynn analysed thousands of phylogenetic trees derived from the comparison of DNA similarity data from thousands of microorganisms to try to identify the oldest genes and when they might have evolved, and to understand how genes move between organisms to shed light on the nature of LUCA. Their careful analysis showed that early in life’s history, different gene types changed at different rates. This suggests that early mutation rates were much higher than at present and there has been a significant contribution of ‘gene jumping’ over time which makes a simple interpretation of the early ‘family tree’ of life misleading. They concluded that previous studies sometimes vastly under-sampled the available data and that the data cannot resolve these questions, but that it does show that early evolution was wildly different from what it is at present.
Professor McGlynn explains, ‘A fundamental question in biology is what were the first life forms on Earth. There are two basic ways to try and address this. First, we can use the comparison of gene sequences to try and understand which ones seem most ancient. Second, we can look for evidence biology may have left in the geological record.’ McGlynn says this work shows that although it is clear there is a fuzzy yet remarkable general outline of a family tree of life in the available DNA sequence data, there has been so much evolutionary change that it is still as of yet impossible to say how the earliest organisms made their living or in what types of environments they lived. This is because the signal is simply too noisy due to this early genetic scrambling. As a result, we are still a long way from understanding what the most primitive organisms on Earth were like or the sorts of environments they lived in.
Importantly, however, this study marks the first time scientists have been able to say something about the pace of early evolution. This work shows there is a detectable signal of very rapid early evolution, thus, while we may not know exactly what early organisms were like, it seems likely life was mutating and evolving very quickly early on. Nevertheless, McGlynn believes it is still amazing that this limited information can be understood at all, that it still tells us important things about the evolution of life on Earth, and suggests we need to develop new ways of looking at available DNA data to find novel techniques of learning what Earth’s earliest life was like.
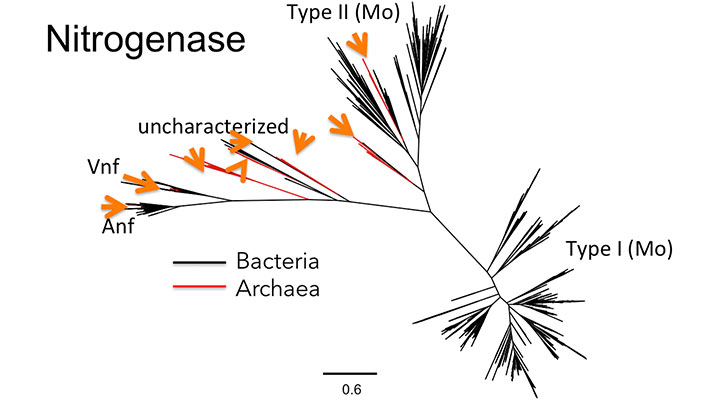
- Figure. 2
- Phylogenetic tree of a protein which is evolutionarily ‘mixed’ between the archaeal and bacterial domains, precluding assignment in the LUCA. Other proteins separate the domains onto different branches, suggesting they are ancient.
Credit: Berkemer and McGlynn 2020. CC BY 4.0
- Reference
| Authors : | Sarah J. Berkemer1,2,3 and Shawn E. McGlynn *4,5,6 |
|---|---|
| Title of original paper : | A new analysis of archaea-bacteria domain separation: variable phylogenetic distance and the tempo of early evolution |
| Journal : | Molecular Biology and Evolution |
| DOI : | 10.1093/molbev/msaa089 |
| Affiliations : | 1 Max Planck Institute for Mathematics in the Sciences, Leipzig, Germany 2 Bioinformatics Group, Department of Computer Science, University Leipzig, Leipzig, Germany 3 Competence Center for Scalable Data Services and Solutions, Dresden/Leipzig, Germany 4 Earth-Life Science Institute, Tokyo Institute of Technology, Meguro, Tokyo, Japan 5 Blue Marble Space Institute of Science, Seattle, WA 6 RIKEN Center for Sustainable Resource Science (CSRS), Saitama Japan |
- ELSI Research Suggests Life Thrived on Earth 3.5 Billion Years Ago | Tokyo Tech News
- Shawn McGlynn - Bridging disciplines and continents | Research Stories | Research
- McGlynn, Shawn | Earth-Life Science Institute
- Researcher Profile | Tokyo Tech STAR Search - Shawn McGlynn
- Earth-Life Science Institute
- Max Planck Institute for Mathematics in the Sciences
- Bioinformatics Group, Department of Computer Science, University Leipzig
- Competence Center for Scalable Data Services and Solutions
- Blue Marble Space Institute of Science
- RIKEN Center for Sustainable Resource Science
- Latest Research News
Further Information
Shawn McGlynn
Associate Professor
Earth-Life Science Institute (ELSI),
Tokyo Institute of Technology
Email mcglynn@elsi.jp
Tel +81-3-5734-3154