Life Science and Technology News
【Labs spotlight】 Junji Hirota Laboratory
The Department has a variety of laboratories for Life Science and Technology, in which cutting-edge innovative research is being undertaken not only in basic science and engineering but also in the areas of medicine, pharmacy, agriculture, and multidisciplinary sciences.
This "Spotlight" series features a laboratory from the Department and introduces you to the laboratory's research projects and outcomes. This time we focus on Junji Hirota Laboratory.

Areas of Supervision
Primary/Life Science and Technology
Professor Junji Hirota![]()
| Degree | PhD 1995, Tokyo Institute of Technology |
|---|---|
| Areas of Research | Molecular Neuroscience, Sensory Systems, Genome Engineering |
| Keywords | Olfactory system, Olfaction, Transcription factor, Bio-imaging, Genome Editing, Synthetic Genome |
| Web site |
Research interest
Olfaction, the sense of smell, is essential for the survival of individuals and species. Odorant receptors (ORs) have evolved to adapt to their chemical environments and this has driven the establishment and diversification of the largest gene family in vertebrates, i.e., ~1,100 functional OR genes in mice, and ~400 in humans. Phylogenetic analyses separate mammalian OR sequences into two distinct groups, which are referred to as class I and class II ORs. Class I OR genes, which were first identified in fish and then in frog, were initially considered to be evolutionary relics in mammals. However, genomic analyses have revealed that mammals possess a substantial number of functional class I OR genes, suggesting that class I ORs have physiologically important functions. On the other hand, class II OR genes are specific to terrestrial animals and account for ~90% of the mammalian OR repertoire. Intriguingly, individual olfactory sensory neurons (OSNs) are believed to express a single functional allele of a single OR gene from the repertoire of either class I or class II ORs.
Our research team aims to understand the molecular mechanisms underlying the binary fate specification of OSNs between two classes of ORs and the subsequent singular OR gene choice. In addition, we also work on the development and application of the Bacillus subtilis genome (BMG) vector system to mouse genome engineering.
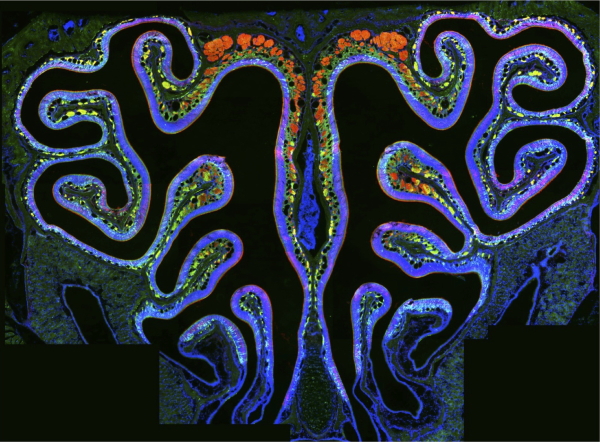
Figure 1. The mouse main olfactory epithelium is a complex mosaic of ~2,200 OSN populations, each defined as expressing one allele from a choice of 2 x ~1,100 OR genes.
1. Molecular mechanism of the singular OR gene expression
For the singular OR gene expression, OSNs must first choose an enhancer element. However, we have an incomplete picture of OR enhancers. In fact, only a few enhancer elements have been identified. Moreover, the repertoire of OR is classified into two groups, referred to as class I and class II ORs, but none of enhancers for class I are identified to date. Because class I OR genes form one of the largest gene clusters in the mouse genome, i.e., 158 class I genes reside in a single cluster spread over a ~3 megabase genomic region, it has been challenging to manipulate such large genomic region to identify enhancer elements.
Recently, by using a combination of a novel genome engineering technique of the Bacillus subtilis genome vector system, bioinformatics, and the CRISPR/Cas9 system, we, for the first time, identified an enhancer element for mouse class I OR genes, named the J element, which exhibits a unique feature with respect to its extraordinary long-range regulation. The J element regulates class I gene expression of a much larger number of genes (75 OR genes) and over a much greater genomic distance (~3 Mb) than class II gene enhancers (7 - 10 genes, ~200 kb). Moreover, its extraordinary long-range regulation surpasses any other known enhancer elements. In addition, our data, which revealed the evolutionarily conserved OR enhancer element across mammalian species shed insights into a long lasting mystery in OR evolution why the class I genes stay in a single cluster on a single chromosome during evolution (whereas class II genes spread over almost all chromosomes by duplications and translocations). We also provide genetic evidence that the allelic exclusion of OR gene is determined by the activity of the J element, and thus propose a new model of the enhancer-dependent singular OR gene expression.
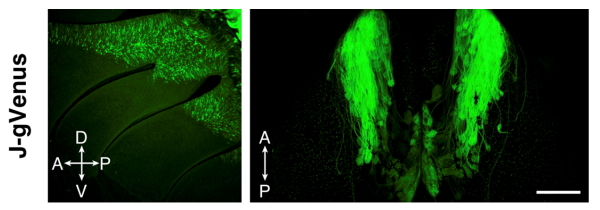
Figure 2. The J-element, a newly identified enhancer element for class I OR genes activate Venus expression specifically in class I OSNs. (Iwata et al, Nature Communications, 2017)
2. Molecular mechanism of the binary fate specification of olfactory sensory neurons.
In the quarter-century since odorant receptor (OR) genes were first identified (Buck and Axel, Cell 1991), genome analyses have uncovered a complete picture of the huge repertoire of OR genes in various species. Each olfactory sensory neuron expresses a single OR gene from either the class I or class II repertoire to give rise two distinct olfactory sensory neuron populations, a fundamental distinction that is critical to both the anatomical and functional organization of olfactory system However, while we have known that two classes of ORs exist for more than 20 years, the mechanisms that regulate the class choice of olfactory sensory neurons have remained an open question. By screening OR-class specific transcription factors and a series of mouse genetic experiments, we study on the molecular mechanisms underlying the binary fate specification of OSNs.
3. Chemosensory cells in various organs.
The transient receptor potential M5 (Trpm5) was first identified in sweet, bitter, and umami taste cells, and plays a critical role in taste signaling. Interestingly, Trpm5-expressing cells have been also found in several extraoral tissues, and their function as chemosensory cells have gradually emerged. For example, in the respiratory epithelium and the urethral epithelium, Trpm5-expressing cells express taste receptors and their downstream signaling molecules, and respond to classical bitter substances and bacterial signaling molecules, inducing physiological protective responses. In the intestinal epithelium, Trpm5-expressing tuft cells release interleukin-25 to initiate type II immune response and clear the gut from the parasites, further providing experimental evidences that Trpm5-expressing cells function as a gatekeeper of biophylactic reactions. We study on the molecular mechanisms of generation of Trpm5-expressing chemosensory cells and their physiological functions.
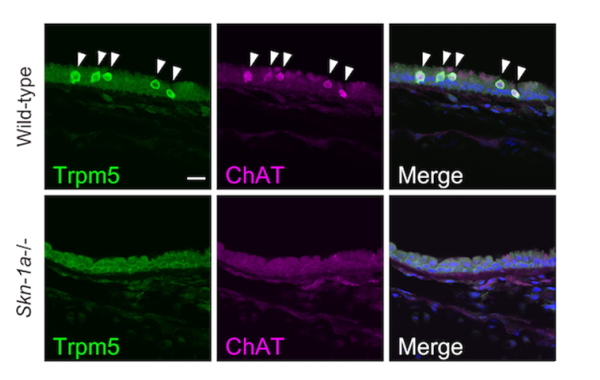
Figure 3. Immunofluorescent images of trachea in wild-type (top row) and Skn-1a knockout mice (bottom row).
Immunosignals of Trpm5 and choline acetyltransferase (ChAT) observed in wild-type mice were no longer observed in Skn-1a-deficient mice, indicating that Skn-1a is essential for the functional differentiation of Trpm5-positive tracheal brush cells. (Yamashita, et al., PLOS ONE 2017).
4. The Bacillus subtilis genome vector system.
The Bacillus subtilis genome (BGM) vector is a novel cloning system for large DNA fragments developed by Prof. Itaya at Keio Univ., in which the entire 4.2 Mb genome of B. subtilis functions as a vector. The BGM vector system has several attractive properties, such as a large cloning capacity of over 3 Mb, stable propagation of cloned DNA and various modification strategies using RecA-mediated homologous recombination. However, genetic modifications using the BGM vector system have not been fully established, and this system had not been applied to transgenesis. In our laboratory, we have applied the BGM system to mouse genome, and have developed the genetic modification methods of the BGM vector system and the BGM transgenesis techniques.
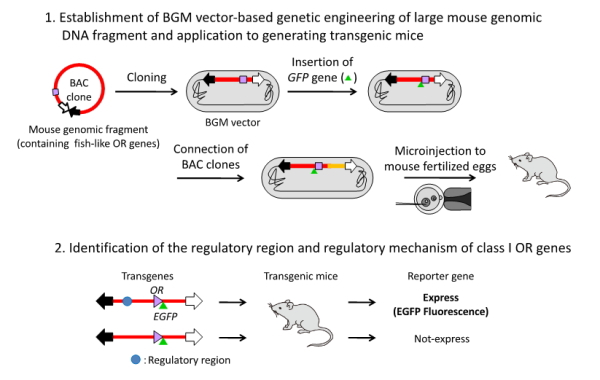
Figure 4. Schematic diagram of the application of the BGM system to mouse genome to identify an enhancer element for class I OR genes.
Publications
- [1] Skn-1a/Pou2f3 functions as a master regulator to generate Trpm5-expressing chemosensory cells in mice. Junpei Yamashita, Makoto Ohmoto, Tatsuya Yamaguchi, Ichiro Matsumoto and Junji Hirota*, PLOS ONE (2017) 12 (12): e0189340, DOI: 10.1371/journal.pone.0189340
- [2] A Long-Range cis-Regulatory Element for Class I Odorant Receptor Genes. Tetsuo Iwata, Yoshihito Niimura, Chizuru Kobayashi, Daichi Shirakawa, Hikoyu Suzuki, Takayuki Enomoto, Kazushige Touhara, Yoshihiro Yoshihara and Junji Hirota*, Nature Communications (2017) 8 (1): 885, DOI: 10.1038/s41467-017-00870-4
- [3] Genetic Lineage Tracing in Taste Tissues Using Sox2-CreERT2 Strain. Makoto Ohmoto, Wenwen Ren, Yugo Nishiguchi, Junji Hirota, Peihua Jiang and Ichiro Matsumoto*, Chemical Senses (2017) 42 (7), 547-552
- [4] Bcl11b/Ctip2 is required for development of lingual papillae in mice. Yugo Nishiguchi, Makoto Ohmoto, Jun Koki, Takayuki Enomoto, Ryo Kominami, Ichiro Matsumoto and Junji Hirota*, Developmental Biology (2016) 416 (1), 98-110
- [5] An inducible recA expression Bacillus subtilis genome vector for stable manipulation of large DNA fragments. Takafumi Ogawa, Tetsuo Iwata, Shinya Kaneko, Mitsuhiro Itaya and Junji Hirota*, BMC Genomics (2015) 16:209, DOI: 10.1186/s12864-015-1425-4
- [6] Skn-1a/Pou2f3 is required for the generation of Trpm5-expressing microvillous cells in the mouse main olfactory epithelium. Tatsuya Yamaguchi, Junpei Yamashita, Makoto Ohmoto, Imad Aoud?, Tatsuya Ogura, Wangmei Luo, Alexander A Bachmanov, Weihong Lin, Ichiro Matsumoto and Junji Hirota*, BMC Neuroscience (2014), 15:13, DOI: 10.1186/1471-2202-15-13 (Recommended by Faculty of 1000)
- [7] Bacillus subtilis genome vector-based complete manipulation and reconstruction of genomic DNA for mouse transgenesis. Tetsuo Iwata, Shinya Kaneko, Yuh Shiwa, Takayuki Enomoto, Hirofumi Yoshikawa and Junji Hirota*, BMC Genomics (2013) 14:300, DOI:10.1186/1471-2164-14-300
- [8] Bcl11b/Ctip2 controls the differentiation of vomeronasal sensory neurons in mice. Takayuki Enomoto, Makoto Ohmoto, Tetsuo Iwata, Ayako Uno, Masato Saitou, Tatsuya Yamaguchi, Ryo Kominami, Ichiro Matsumoto and Junji Hirota*, Journal of Neuroscience (2011), 31(28), 10159-10173
- [9] Differential impact of Lhx2 deficiency on expression of class I and class II odorant receptor genes in mouse. Junji Hirota, Masayo Omura and Peter Mombaerts*, Molecular and Cellular Neuroscience (2007), 34, 679-688
- [10] The promoter of the mouse odorant receptor gene M71. Andrea Rothman, Paul Feinstein, Junji Hirota and Peter Mombaerts*, Molecular and Cellular Neuroscience (2005), 28, 535-548
- [11] The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Junji Hirota and Peter Mombaerts*, Proceedings of the National Academy of Science, USA (2004), 101, 8751-8755
- [12] Targeting a complex transcriptome: The construction of the mouse full-length cDNA encyclopedia. Piero Carninci, Kazunori Waki, Toshiyuki Shiraki, Hideaki Konno, Kazuhiro Shibata, Masayoshi Itoh, Katsunori Aizawa, Takahiro Arakawa, Yoshiyuki Ishii, Daisuke Sasaki, Hidemasa Bono, Shinji Kondo, Yuichi Sugahara, Rintaro Saito, Naoki Osato, Shiro Fukuda, Kenjiro Sato, Akira Watahiki, Tomoko Hirozane-Kishikawa, Mari Nakamura, Yuko Shibata, Ayako Yasunishi, Noriko Kikuchi, Atsushi Yoshiki, Moriaki Kusakabe, Stefano Gustincich, Kirk Beisel, William Pavan, Vassilis Aidinis, Akira Nakagawara, William A. Held, Hiroo Iwata, Tomohiro Kono, Hiromitsu Nakauchi, Paul Lyons, Christine Wells, David A. Hume, Michela Fagiolini, Takao K. Hensch, Michelle Brinkmeier, Sally Camper, Junji Hirota, Peter Mombaerts, Masami Muramatsu, Yasushi Okazaki, Jun Kawai and Yoshihide Hayashizaki*, Genome Research (2003), 13(6B), 1273-1289 (Recommended by Faculty of 1000)
- [13] Combinatorial coexpression of neural and immune multigene families in mouse vomeronasal sensory neurons. Tomohiro Ishii, Junji Hirota and Peter Mombaerts*, Current Biology, (2003), 13, 394-400
- A whole-body approach to understanding chemosensory cells | Life Science and Technology News
- Pioneering Discovery of an Odor-detecting Receptor Enhancer | Life Science and Technology News
Contact
Professor Junji Hirota
Room 201, B1B2-Annex C, Suzukakedai campus
E-mail : jhirota@bio.titech.ac.jp
*May 1, 2025:Some of the content has been updated with the latest information.