Life Science and Technology News
【Labs spotlight】Kawai Laboratory
The Department has a variety of laboratories for Life Science and Technology, in which cutting-edge innovative research is being undertaken not only in basic science and engineering but also in the areas of medicine, pharmacy, agriculture, and multidisciplinary sciences.
This "Spotlight" series features a laboratory from the Department and introduces you to the laboratory's research projects and outcomes. This time we focus on Kawai Laboratory working on the development of single molecule analysis and diagnosis methods.

Areas of Supervision
Primary/Life Science and Technology
Professor Kiyohiko Kawai![]()
| Office | Room 1131, B2 building, Suzukakedai campus (till 2023.3 Room 821) |
|---|---|
| Degree | PhD 1999, Kyoto University |
| Areas of Research | Bioorganic chemistry, photochemistry |
| Keywords | Fluorescence, Blinking, Single molecule detection, Nucleic Acids |
| WEBsite URL | Kawai Lab. |
Research interest
Investigation of various biological dynamic phenomena based on the development of single-molecule level analytical methods
We are interested in biological phenomena taking place in a relatively short time range (microsecond to second (10-6 - 100 sec.)). To access such phenomena, conventional approaches such as transient absorption measurements or NMR usually require substantial amount of sample (> 1 nmol). In order to achieve ultralow detection limit, one strategy would be to focus on a detection method that relies on the properties of molecules that become highlighted when we look at molecules at the single-molecule level. Among such phenomena, we have focused on the fluctuating emissions between bright “ON” and dark “OFF” states of fluorescent molecules, so-called “blinking”. During the repetitive cycles of excitation and emission, fluorescent molecules may occasionally enter non-fluorescent off states, such as a triplet state, a radical ion state, and an isomerized state. Reversible formation of such OFF states causes a blinking of the fluorescence. By measuring the duration of the ON time (τON) and OFF time (τOFF) of the blinking, we can investigate various biological phenomena with sub-microsecond time resolution at the single-molecule level. The changes in the surrounding local microenvironment that modulate blinking would also be approached at the single-molecule level by Kinetic Analysis based on the Control of the fluorescence Blinking (KACB method).
Reviews: Accounts Chem. Res., 54, 1001-1010 (2021).![]() Chem. Eur. J., 26, 7740-7746 (2020).
Chem. Eur. J., 26, 7740-7746 (2020).![]()
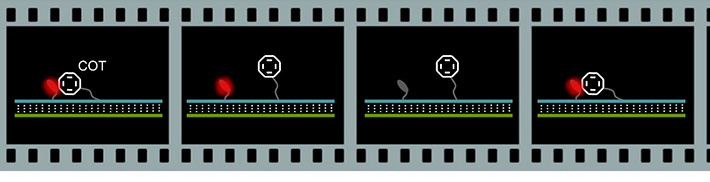
・Fluorescent blinking triggered by Triplet-Triplet energy transfer (TTET)
Cyclooctatetraene (COT) was used as both a triplet acceptor and a photo-stabilizing agent to control and observe the blinking at the single-molecule level. By using DNA as a platform, we demonstrated that triplet blinking of the fluorescent molecule ATTO 647N can be controlled by the collision reaction between COT. Dynamics of biomolecules can be investigated by measuring the blinking. COT and ATTO 647N were attached to the molecular beacon type probe which allowed us the detection of model biomarker miR-155 at the single-molecule level.
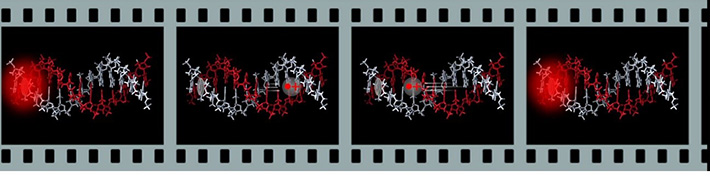
・Fluorescent blinking triggered by charge transfer in DNA
By utilizing the transient absorption measurements, we previously showed that charge transfer dynamics in DNA is strongly affected by the DNA sequence. We showed that DNA sequence information including the data on single nucleotide polymorphisms (SNPs) can be read-out by measuring the charge transfer kinetics. However, the measurement requires a significant amount of sample (>1 nmol) and thus cannot be used for diagnosis. We focus on that, by using a fluorophore as a photosensitizer to generate charges on DNA, the charge-separation, charge-transfer, and charge-recombination dynamics in DNA can be monitored as the fluorescence blinking. In this case, the blinking was caused by the successive reduction and re-oxidation cycles of the fluorescent molecule ATTO 655. The τOFF value corresponds to the lifetime of the charge-separated state. Based on the fact that the charge-transfer dynamics in DNA is strongly affected by the DNA sequence, we demonstrated that single-nucleotide differences in DNA that modulate the charge-recombination kinetics can be detected by monitoring the blinking of the fluorescence. We succeeded in in situ detection of mRNA point mutation on pathological section.
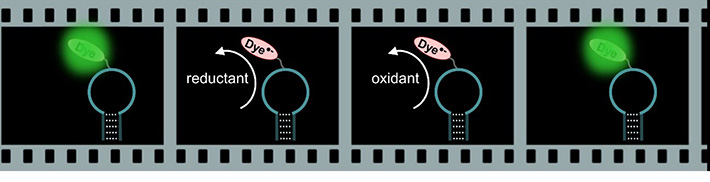
・Fluorescent blinking triggered by redox reaction
We focused on the control of redox blinking to monitor the microenvironment of the fluorescent molecule. By adding ascorbic acid 2-phosphate (VcP) as a reductant, the triplet state of fluorescent molecule was converted to the radical anion OFF-state. The duration of the OFF state corresponds to the lifetime of the radical anion of fluorescent molecule. Here, a bulky oxidant, diethylenetriaminepentaacetic acid iron(III) (FeDTPA), was utilized to regenerate the intact fluorescent molecule at the ground state. Owing to the larger size of FeDTPA, the bimolecular reaction rate between the radical anion of fluorescent molecule and FeDTPA changed dramatically along with the changes in microenvironment of fluorescent molecule. Observation of the redox blinking enabled sensitive detection of subtle conformational changes around the fluorescent molecule caused by single nucleotide alternations in the DNA sequence. Single molecule detection of target DNA was achieved.
・Fluorescent blinking triggered by trans-cis isomerization
Cyanine dyes such as Cy3 are widely used as a fluorescent probe to investigate various biological phenomena. It is known to undergo trans-cis photo-isomerization and successive cis-trans back thermal-isomerization by rotation around the C–C bonds of the poly-methine chain. This cis-trans isomerization causes the blinking of the fluorescence. We focus on the fact that the size of the Cy3 is just about the same as the width of the triple helix. Since the cis-trans isomerization efficiency is considered to be strongly dependent on the steric effects that impact the rotation of the molecule, we hypothesized that Cy3 may exhibit a triple helix-specific blinking. We incorporated Cy3 into DNA double helix, or triple helix conformation and investigated the differences in blinking behavior by Fluorescence Correlation Spectroscopy (FCS). The duration of the on time (τON) and off time (τOFF) of the blinking, which reflect the inverse of the trans-cis photo-isomerization rate and the cis-trans back isomerization rate, respectively, were highest in the triple helix conformation. These results suggest that Cy3 can be used to track the presence of the triple helix conformation at the single molecule level.
Selected publications
Shuya Fan, Tadao Takada, Atsushi Maruyama, Mamoru Fujitsuka, Kiyohiko Kawai*
Programmed Control of Fluorescence Blinking Patterns Based on Electron Transfer in DNA
Chem. Eur. J. DOI: 10.1002/chem.202203552![]()
Shuya Fan, Tadao Takada, Atsushi Maruyama, Mamoru Fujitsuka, Kiyohiko Kawai*
Large Heterogeneity Observed in Single Molecule Measurements of Intramolecular Electron Transfer Rates through DNA
Bull. Chem. Soc. J. 95, 1697-1702 (2022)![]()
Shuya Fan, Jie Xu, Yasuko Osakada, Katsunori Hashimoto, Kazuya Takayama, Atsushi Natsume, Masaki Hirano, Atsushi Maruyama, Mamoru Fujitsuka, Kumi Kawai*, Kiyohiko Kawai*
Electron-Transfer Kinetics through Nucleic Acids Untangled by Single-Molecular Fluorescence Blinking
Chem 8, 3109-3119 (2022)![]() Press release
Press release![]()
Kiyohiko Kawai*, Mamoru Fujitsuka
Single-Molecule Fluorescence Kinetic Sandwich Assay Using a DNA Sequencer
Chem. Lett. 51, 139-141 (2022)![]()
Jie Xu, Shuya Fan, Lei Xu, Atsushi Maruyama, Mamoru Fujitsuka, Kiyohiko Kawai*
Control of Triplet Blinking Using Cyclooctatetraene to Access the Dynamics of Biomolecules at the Single‐Molecule Level
Angew. Chem. Int. Ed.60, 12941-12948 (2021)![]() Press release
Press release![]()
Kiyohiko Kawai*, Mamoru Fujitsuka, Atsushi Maruyama
Single-Molecule Study of Redox Reaction Kinetics by Observing Fluorescence Blinking,
Acc. Chem. Res. 54, 1001-1010 (2021)![]()
Contact
Professor Kiyohiko Kawai
Room 1131, B2 building, Suzukakedai campus (till 2023.3 Room 821)
E-mail : kawai.k@bio.titech.ac.jp
Tel / Fax : +8145-924-5821
*Find more about the lab and the latest activities at the lab site![]() .
.
*May 1, 2025:Some of the content has been updated with the latest information.