Life Science and Technology News
【Labs spotlight】Osakabe Laboratory
The Department has a variety of laboratories for Life Science and Technology, in which cutting-edge innovative research is being undertaken not only in basic science and engineering but also in the areas of medicine, pharmacy, agriculture, and multidisciplinary sciences. This "Spotlight" series features a laboratory from the Department and introduces you to the laboratory's research projects and outcomes. This time we focus on Prof. Yuriko Osakabe’s Laboratory.

Areas of Supervision
Primary/Life Science and Technology
Secondary/Science and Technology for health Care and Medicine
Professor Yuriko Osakabe![]()
※ The laboratory website was released on November 21, 2022.
| Degree | PhD 1996, Tokyo University of Agriculture and Technology |
|---|---|
| Areas of Research | Molecular biology, Plant biology, Synthetic biology |
| Keywords | genome editing, genetic engineering, plant, stress response |
| Web site |
Research interest
Our laboratory conducts fundamental research on genome editing, which is involved in the modification of various biological functions, and also promotes research on molecular mechanisms of environmental stress responses in plants, especially on the molecular breeding of plants in response to environmental changes.
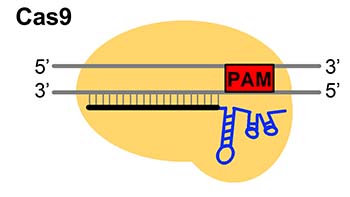
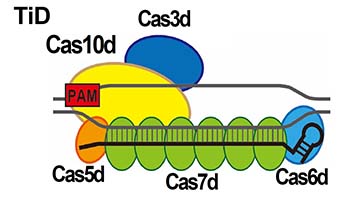
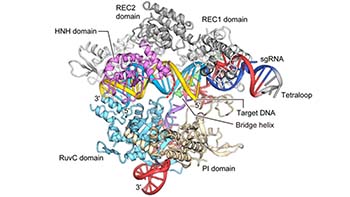
(Modified from Nishimasu et al. 2014 Cell)
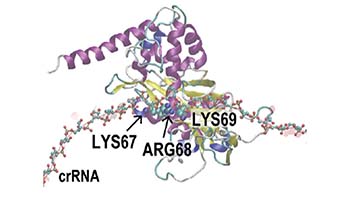
of TiD (Cas7d) -sgRNA-DNA complex
(Osakabe et al. 2021 Nucleic Acid Research)
Fig.1 Genome editing technologies CRSIPR-Cas9 and CRISPR-Cas type I-D, “TiD”.
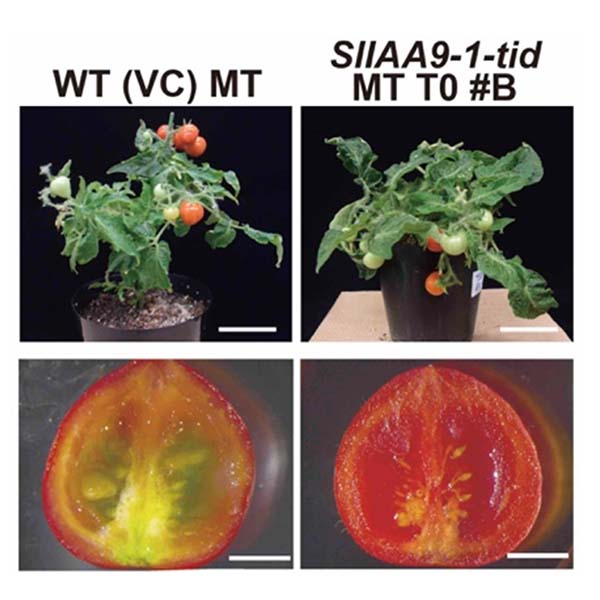
Fig.2. Generation of the SlIAA9 tomato mutant by genome editing.
Genome editing targeting the tomato IAA9 gene to produce mutants that exhibit
parthenocarpy (a trait that allows fruit set without artificial pollination)
(from Osakabe et al., 2020, Comm. Biol.).
Genome editing is a technology that modifies gene functions by introducing target sequence-specific mutations into DNA/RNAs of a wide range of organisms, from microorganisms to higher eukaryotes, through the use of artificial nucleases that specifically digest the target sequences. In recent years, the applied research of genome editing has made great progress, and it is becoming widely used in a variety of fields, such as medical and drug discovery, gene and cell therapy, to overcome intractable diseases, various industries that utilize biological resources, and development of new useful breeds in agriculture. CRISPR-Cas9, which is widely used for genome editing, is one of the acquired immune systems of microorganisms. It is composed of Cas9, which functions for target DNA cleavage, and a 20-base RNA molecule (RNA complementary to the target DNA), which has an role on recognition of the target DNA sequence. Among the various CRISPR-Cas in microorganisms, we focused on CRISPR-Cas type I-D in the cyanobacterium, Microcystis aeruginosa, whose function had been unknown. We have named this system "TiD" and are conducting detailed studies to elucidate the mutation patterns in plant and animal cells and to improve the efficiency of mutagenesis.
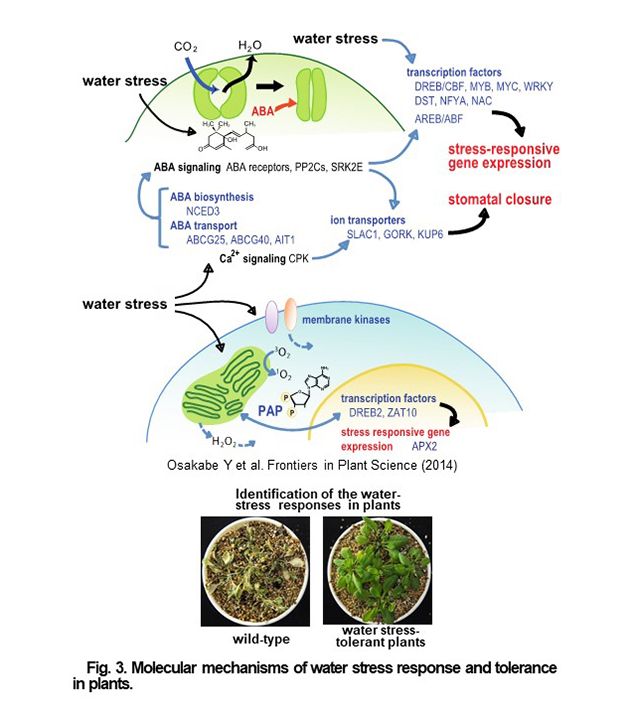
Since plants cannot move, the sensing the external environment is important for their life support, and water in particular has a significant impact on plant survival and productivity. In particular, water has a significant impact on plant survival and productivity. Plants have developed the response to environment during evolution. Stomatal opening and closing, in particular, regulates the balance between water stress and photosynthesis.
In the sensing of water deficit stress and downstream pathways of the signal transduction, a variety of regulatory processes such as biosynthesis of plant hormones, activation of transcription factors and membrane- localized transporters, and regulation of important metabolic pathways control and function in response to environmental conditions to flexibly respond and survive to adverse environmental conditions. We are investigating the novel roles of key functional proteins in plant environmental responses, with the aim of elucidating the flexible and sophisticated survival strategies of plants in response to stressful environments. Through new genetic engineering technologies, such as genome editing, we are conducting applied researches to apply the knowledge gained from our studies on the molecular mechanisms of environmental responses to molecular breeding of crops. We expect to be able to produce the novel useful crops that are tolerant to environmental stress.
Dr. Yuriko Osakabe’s Laboratory is a new laboratory started in April 2021. Through our basic research on genome editing technology and the molecular mechanisms of plant responses to the environment, as well as the applied field of genetic engineering, molecular breeding and the development of peripheral technologies, we hope that you will experience and study the importance and excitement of life science, which challenges the solution of various social problems such as food crises and the treatment of intractable diseases.
Selected publications
- 1.Osakabe, K., Wada, N., Murakami, E., Miyashita, N., Osakabe, Y. (2021) Genome editing in mammalian cells using the CRISPR type I-D nuclease. Nucleic Acids Res. 49, 6347. doi: 10.1093/nar/gkab348.
- 2.Abe-Hara, C., Yamada, K, Wada, N., Ueta, R., Hashimoto, R., Osakabe, K., Osakabe, Y. * (2021) Effects of the sliaa9 mutation on shoot elongation growth of tomato cultivars. Front Plant Sci. 12, 627832. doi: 10.3389/fpls.2021.627832.
- 3.Osakabe, K.*, Wada, N., Miyaji, T., Murakami, E., Marui K., Ueta, R., Hashimoto, R., Abe-Hara, C., Kong, B., Yano, K., Osakabe, Y.* (2020) Genome editing in plants using CRISPR type I-D nuclease. Communications. Biol. 3, 648. *Corresponding authors.
- 4.Wakabayashi, T., Hamana, M., Mori, A., Akiyama, R., Ueno, K., Osakabe, K., Osakabe, Y., Suzuki, H., Takikawa, H., Mizutani, M., Sugimoto, Y. (2019) Direct conversion of carlactonoic acid to orobanchol by cytochrome P450 CYP722C in strigolactone biosynthesis. Science Advances, 5, eaax9067
- 5.Toda E, Koiso N, Takebayashi A, Ichikawa M, Kiba T, Osakabe K, Osakabe Y, Sakakibara H, Kato N, Okamoto T (2019) An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice. Nature Plants, 5, 363-368.
- 6.Osakabe, Y. *†., Liang, Z†., Ren, C., Nishitani, C., Osakabe, K., Wada, M., Komori, S., Malnoy, M., Velasco, R., Jung, M-H., Koo, O-J., Viola, V., and Kanchiswamy, C.N*†. (2018) CRISPR/Cas9 mediated genome editing in Apple and Grapevine. Nature Protocols, 13, 2844-2863. *Corresponding authors, †Equally contribution
- 7.Takahashi, F., Suzuki, T., Osakabe, Y., Betsuyaku, S., Kondo, Y., Dohmae, N., Fukuda, H., Yamaguchi-Shinozaki, K., Shinozaki, K. (2018) A small peptide modulates stomatal control via abscisic acid in long distance signalling. Nature, 556, 235?238.
- 8.Li, W., Nguyen, K., Chu, H.D., Ha, C.V., Watanabe, Y., Osakabe, Y., Leyva-Gonzalez, M., Sato, M., Toyooka, K., Voges, L., Tanaka, M., Mostofa, M.G., Seki, M., Seo, M., Yamaguchi, S., Nelson, D.C., Tian, C., Herrera-Estrella, L., Tran, L-S (2017) The karrikin receptor KAI2 promotes drought resistance in Arabidopsis thaliana. PLoS Genetics, 13(11): e1007076.
- 9.Nishitani, C., Hirai, N., Komori, S., Wada, M., Okada, K., Osakabe, K., Yamamoto, T., and Osakabe, Y. * (2016) Efficient Genome Editing in Apple Using a CRISPR/Cas9 system. Sci. Reports, 6, 31481, doi:10.1038/srep31481 *Corresponding author
- 10.Osakabe, Y. *., Watanabe, T., Sugano, SS., Ueta, R., Ishihara, R., Shinozaki, K., and Osakabe, K. (2016) Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Reports, 6, 26685. *Corresponding authors
- 11.Osakabe, Y*., Osakabe, K., Shinozaki, K., and Tran, L-S* (2014) Response of plants to water stress. Frontiers in Plant Sci., 5: 86. doi:10.3389/fpls.2014.00086
- 12.Osakabe, Y*., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L-S*. (2014) ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol., 202, 35-49. DOI: 10.1111/nph.12613 *Corresponding authors
- 13.Osakabe, Y., Arinaga, N., Umezawa, T., Katsura, S., Nagamachi, K., Tanaka, H., Ohiraki, H., Yamada, K., Souk, S., Abo, M., Yoshimura, E., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2013) Osmotic stress response and plant growth controlled by the potassium transporters in Arabidopsis. Plant Cell, 25, 609-624.
- 14.Osakabe, K. Osakabe, Y., and Toki, S. (2010) Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc. Natl. Acad. Sci. USA, 107 12034-12039.
- 15.Osakabe, Y., Mizuno, S., Tanaka, H., Maruyama, K., Osakabe K, Todaka, D., Fujita, Y., Kobayashi M, Shinozaki, K., and Yamaguchi-Shinozaki, K. (2010) Overproduction of a membrane-bound receptor-like protein kinase1, RPK1, enhances abiotic stress tolerance of Arabidopsis. J. Biol. Chem., 285, 9190-9201.
- 16.Sakuma, Y., Maruyama, K., Qin, F., Osakabe, Y., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006) Dual function of an Arabidopsis transcription factor DREB2A in water-stress- and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA, 103, 18822-18827.
- 17.Osakabe, Y., Maruyama, K., Seki, M., Satou, M., Shinozaki, K., Yamaguchi-Shinozaki, K. (2005) Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell, 17, 1105-1119.
Contact
Professor Yuriko Osakabe
Room 1011, J2 building, Suzukakedai campus
E-mail : osakabe.y.ab@m.titech.ac.jp
*Find more about the lab and the latest activities at the lab site![]() .
.
*May 1, 2025:Some of the content has been updated with the latest information.