Life Science and Technology News
【Labs spotlight】Matsuura Laboratory

Areas of Supervision
Primary/Earth-Life Science
Secondary/Life Science and Technology
(Earth-Life Science Institute)
Professor Tomoaki Matsuura![]()
| Keywords | artificial cells, biotechnology, directed evolution, membrane proteins |
|---|---|
| Web site | Matsuura Lab (Earth-Life Science Institute) |
Research interest
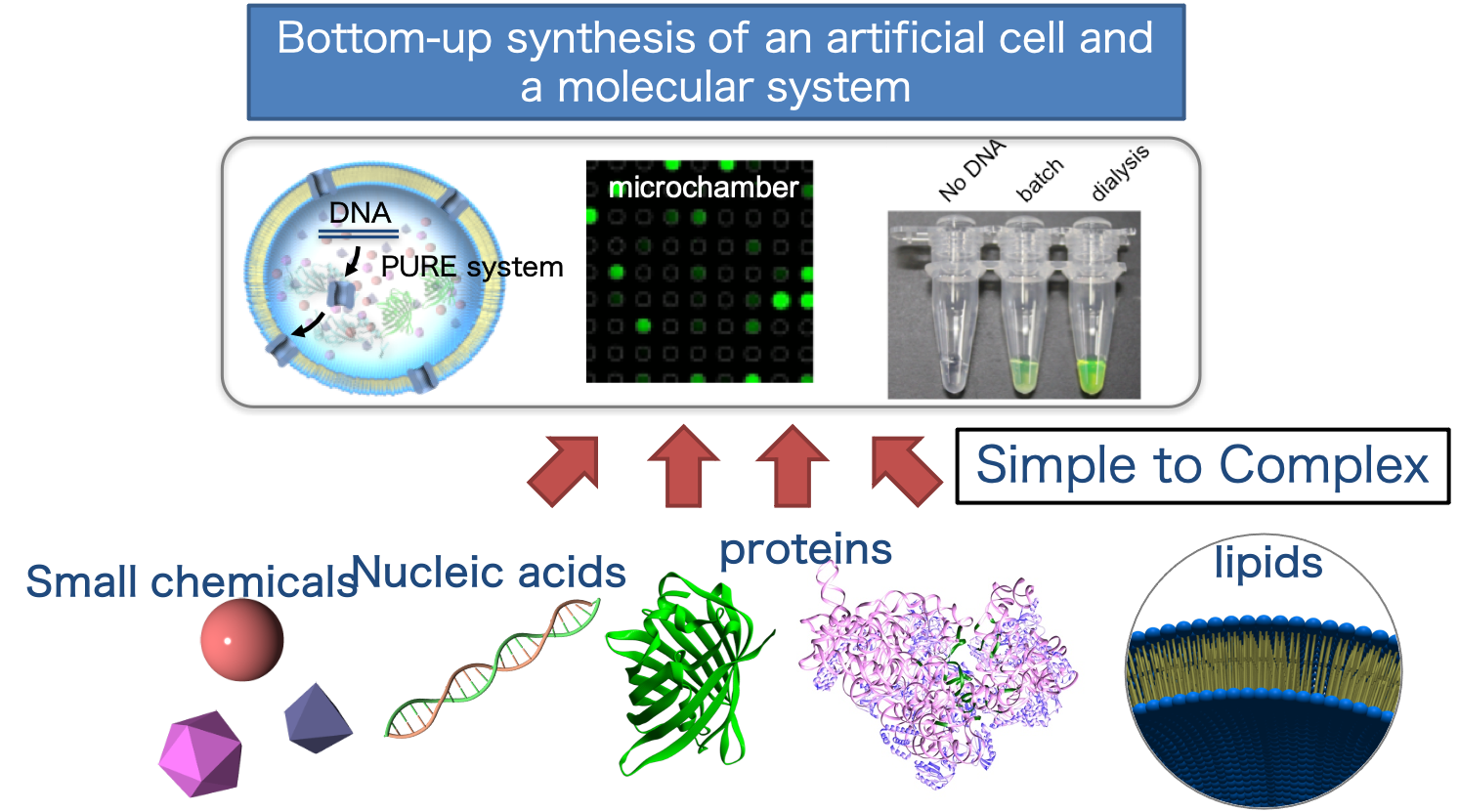
Abstract:Our group combines the components of living organisms to create molecular systems that possess specific features and functions. In this way, we can reveal the properties of primitive cells that may have existed in the early stages of life, or create molecules and molecular systems that contribute to various applications [Figure 1, 2].
Figure 1:Bottom-up synthesis of artificial cells
Figure 2:Artificial cells that look alive
Significance:In recent years, there has been a great interest in the bottom-up construction of molecular systems (e.g., artificial cells) that possess specific functions and properties of cells. This research area can be broadly divided into two areas based on the objective.
(A) Understanding the origin of life and the mechanism of biological phenomena. Even E. coli, a simple model organism, can produce it’s copu in just 20 minutes despite being composed of more than tens of millions of molecules. It also grows and adapts to a variety of environmental conditions by rapidly changing its internal state. It is still unclear how these amazing self-replicating and adaptive abilities can be composed, and how they have emerged. If artificial cells with self-replicable and/or environmental adaptive capacity can be created, the sufficient conditions for building such molecular systems will become clear. In addition, the boundary between non-living and living is expected to be clarified.
(B) Developing application tools including, drug discovery, material production, and chemical detection using artificial cells. Because artificial cells are composed of only defined materials, their composition can be freely adjusted. With this advantage, it is possible to give an artificial cell the desired function. Likewise, it is possible to design them so that they do not have unwanted properties. This is a property that is not present in cells that have an absolute requirement to be "alive".
Research Projects: Research Project.
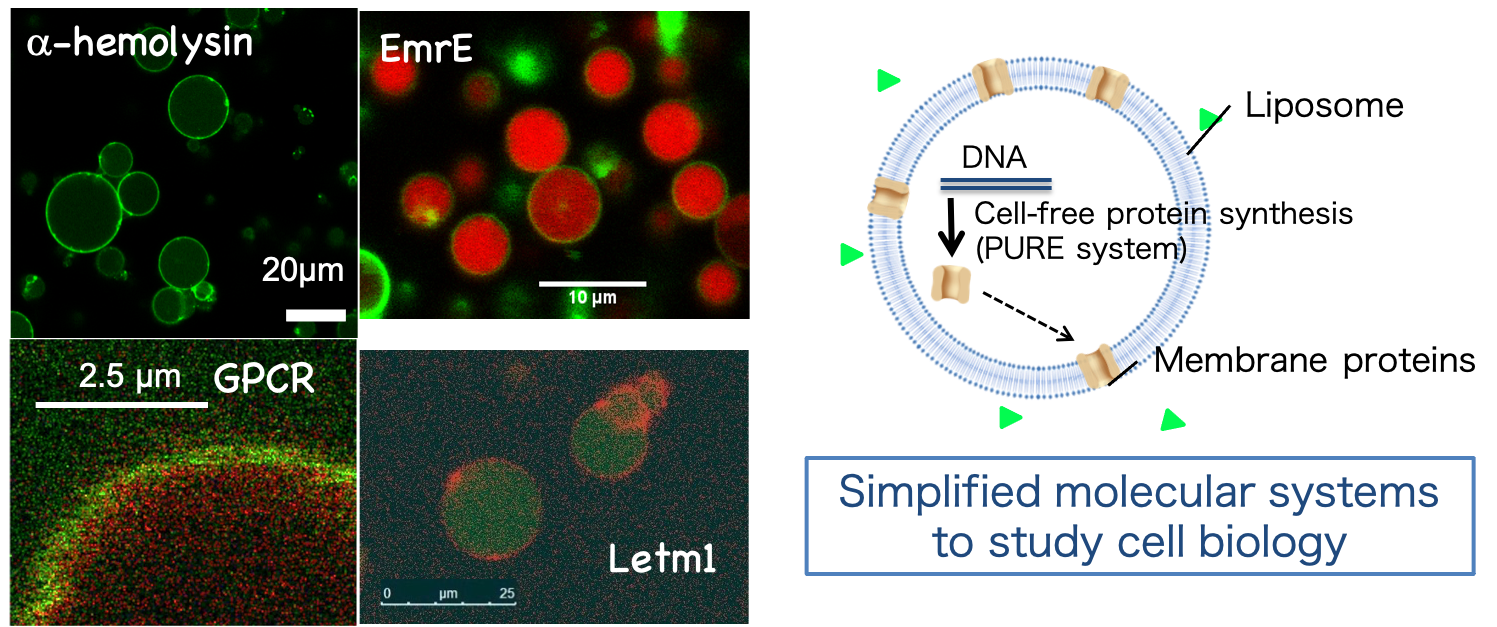
Construction of artificial cells and their applications
Cells have compartments to encapsulate their contents, and inside the cell they synthesize proteins based on the information encoded on the genes. We have created an artificial cell with these features [Figure 3]. These artificial cells are assembled from components. We thus can replace or add desired components to the artificial cells and investigate their properties, thereby investigate the relationship between the components and the properties of the entire system. We also aim to design artificial cells that will contribute to medical applications, biosensors, and material production. For more information on our research, see our website and papers (here!![]() )。
)。
Figure 3: Schematic (right) and micrograph (left) of an artificial cell.
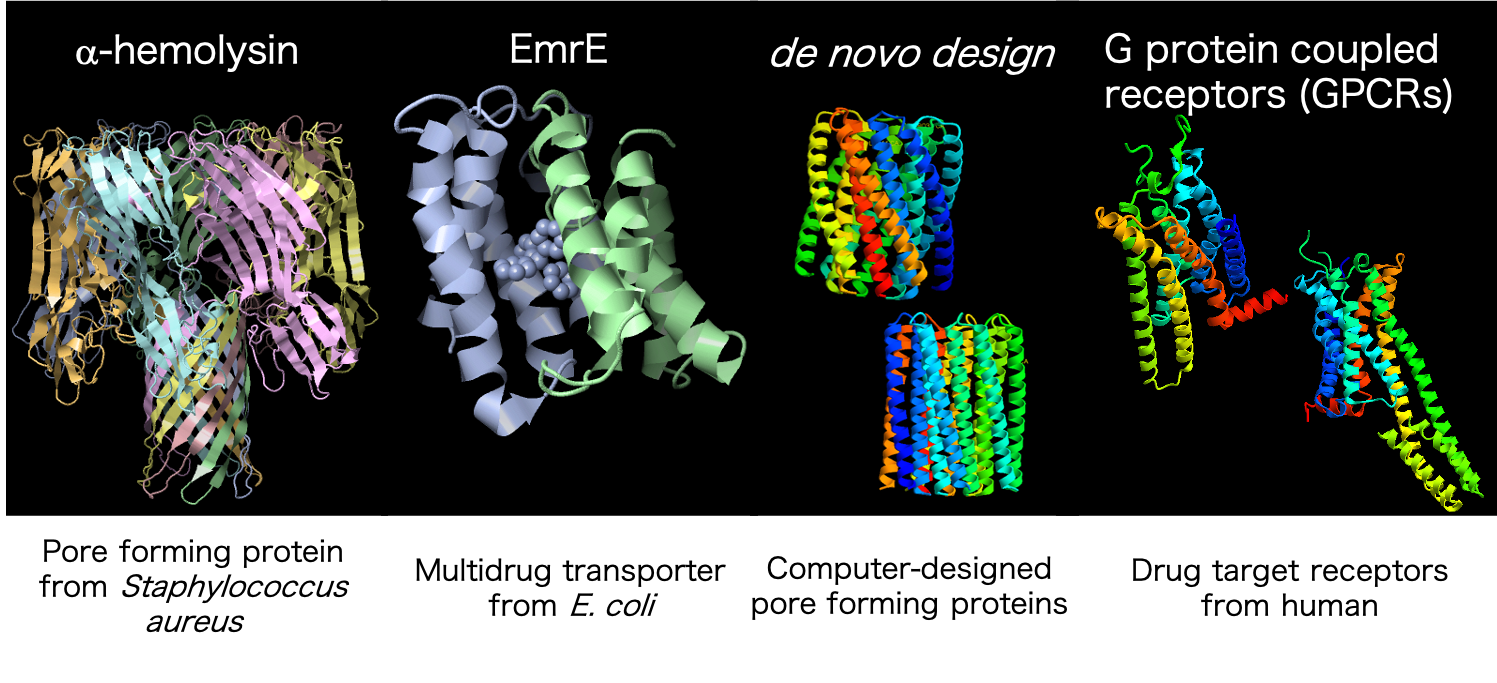
Directed Evolution of Membrane Proteins
Cells have compartments to encapsulate their contents, and have various membrane proteins on the compartment [Figure 4]. Therefore, membrane proteins are one of the most important molecules for cells to detect the external environment with very high sensitivity. They can also transport substances inside and outside the cell across the cell membrane. These properties could be used to make excellent devices for detecting various substances. However, natural membrane proteins are not conveniently made for human use. We are working to synthesize these membrane proteins, evolve their functions in the laboratory, and transform them into human-friendly molecules. For more research, see our website and papers (here!![]() )。
)。
Figure 4: Membrane proteins being evolved in the laboratory. By evolving them, we aim to create molecules for use in biosensors and other applications.
Selected publications
- 1.Xu, C., Lu, P., Gamal El-Din, T. M., Pei, X. Y., Johnson, M. C., Uyeda, A., Bick, M. J., Xu, Q., Jiang, D., Bai, H., Reggiano, G., Hsia, Y., Brunette, T. J., Dou, J., Ma, D., Lynch, E. M., Boyken, S. E., Huang, P. S., Stewart, L., DiMaio, F., Kollman, J. M., Luisi, B. F., Matsuura, T., Catterall, W. A., and Baker, D. (2020) Computational design of transmembrane pores. Nature 585, 129-134
- 2.Noba, K., Ishikawa, M., Uyeda, A., Watanabe, T., Hohsaka, T., Yoshimoto, S., Matsuura, T., and Hori, K. (2019) Bottom-up Creation of an Artificial Cell Covered with the Adhesive Bacterionanofiber Protein AtaA. J Am Chem Soc 141, 19058-19066
- 3.Dwidar, M., Seike, Y., Kobori, S., Whitaker, C., Matsuura, T., and Yokobayashi, Y. (2019) Programmable Artificial Cells Using Histamine-Responsive Synthetic Riboswitch. J Am Chem Soc 141, 11103-11114
- 4.Matsuura, T., Tanimura, N., Hosoda, K., Yomo, T., and Shimizu, Y. (2017) Reaction dynamics analysis of a reconstituted Escherichia coli protein translation system by computational modeling. Proc Natl Acad Sci U S A 114, E1336-E1344
- 5.Soga, H., Fujii, S., Yomo, T., Kato, Y., Watanabe, H., and Matsuura, T. (2014) In vitro membrane protein synthesis inside cell-sized vesicles reveals the dependence of membrane protein integration on vesicle volume. ACS Synth Biol 3, 372-379
- 6.Kazuta, Y., Matsuura, T., Ichihashi, N., and Yomo, T. (2014) Synthesis of milligram quantities of proteins using a reconstituted in vitro protein synthesis system. J Biosci Bioeng 118, 554-557
- 7.Fujii, S., Matsuura, T., Sunami, T., Nishikawa, T., Kazuta, Y., and Yomo, T. (2014) Liposome display for in vitro selection and evolution of membrane proteins. Nat Protoc 9, 1578-1591
- 8.Fujii, S., Matsuura, T., Sunami, T., Kazuta, Y., and Yomo, T. (2013) In vitro evolution of alpha-hemolysin using a liposome display. Proc Natl Acad Sci U S A 110, 16796-16801
- 9.Matsuura, T., Kazuta, Y., Aita, T., Adachi, J., and Yomo, T. (2009) Quantifying epistatic interactions among the components constituting the protein translation system. Mol Syst Biol 5, 297
- 10.*Kazuta, Y., *Adachi, J., *Matsuura, T., Ono, N., Mori, H., and Yomo, T. (2008) Comprehensive analysis of the effects of Escherichia coli ORFs on protein translation reaction. Mol Cell Proteomics 7, 1530-1540 (*contributed equally)
- 11.Matsuura, T., Yamaguchi, M., Ko-Mitamura, E. P., Shima, Y., Urabe, I., and Yomo, T. (2002) Importance of compartment formation for a self-encoding system. Proc Natl Acad Sci U S A 99, 7514-7517
- 12.Matsuura, T., Miyai, K., Trakulnaleamsai, S., Yomo, T., Shima, Y., Miki, S., Yamamoto, K., and Urabe, I. (1999) Evolutionary molecular engineering by random elongation mutagenesis. Nat Biotechnol 17, 58-61
Contact
Professor Tomoaki Matsuura
Room 307, Ishikawadai 7th building , Ookayama campus
E-mail : matsuura_tomoaki@elsi.jp
TEL : 03-5734-2802
*Find more about the lab and the latest activities at the lab site![]()
*May 1, 2025:Some of the content has been updated with the latest information.