Life Science and Technology News
【Labs spotlight】 Fujita Laboratory

Areas of Supervision
Primary/Life Science and Technology
Associate Professor Naonobu Fujita![]()
| Office | Room 210, S2 building, Suzukakedai campus |
|---|---|
| Degree | PhD(Doctor of Engineering) 2009, Waseda University |
| Areas of Research | Cell biology |
| Keywords | Organelle, Muscle, Drosophila, Autophagy, Lysosome |
| Web site | Fujita Lab |
Research interest
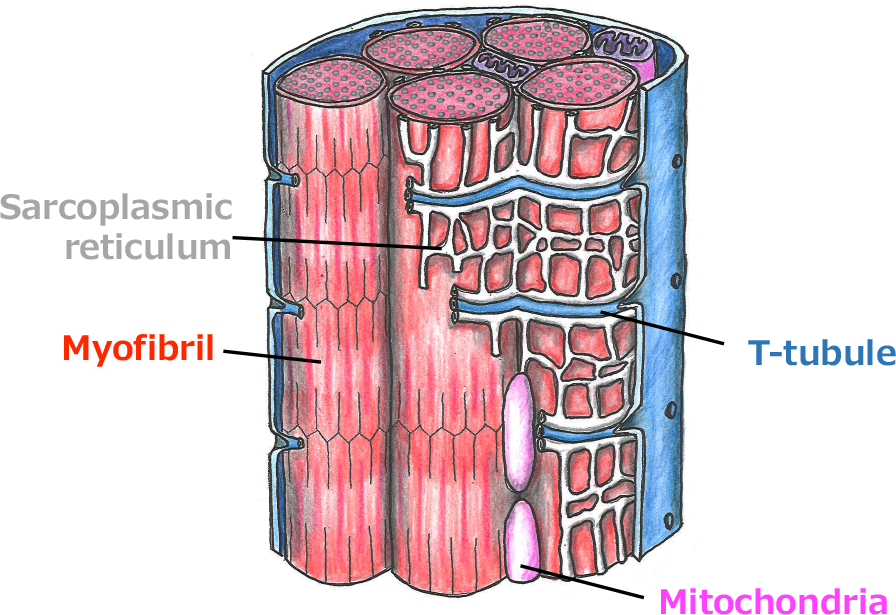
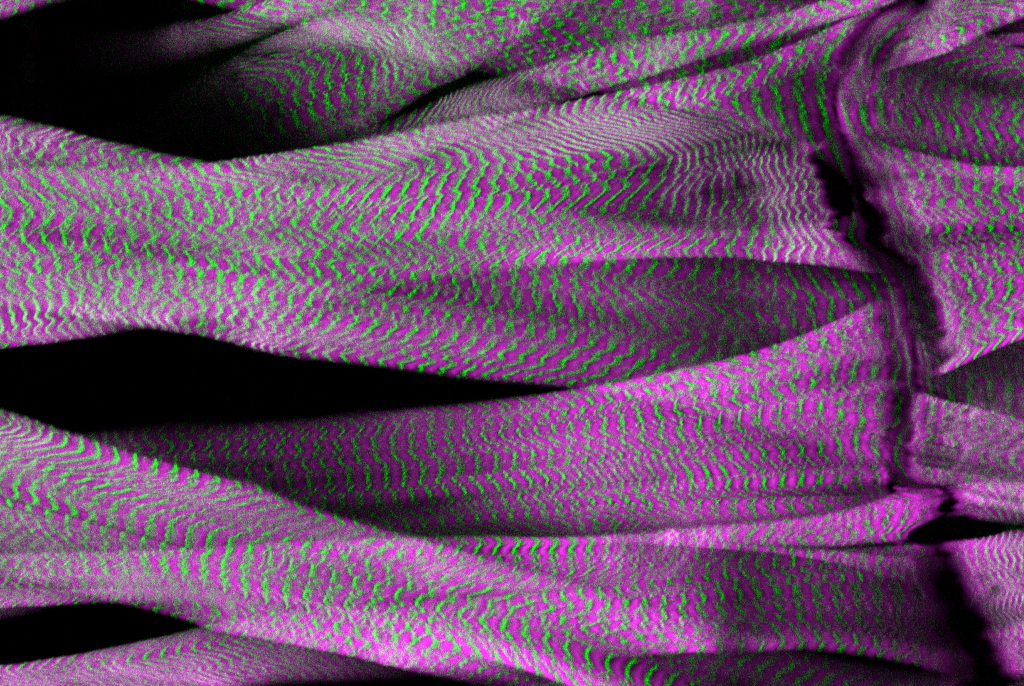
Differentiated muscle cells have highly organized membranous organelles, such as Transverse (T)-tubules required for excitation-contraction coupling. Their roles are established; however, the mechanisms shaping the organelles are largely unknown. Our study aims to elucidate the mechanisms to form and remodel the membranous organelles in muscle cells using Drosophila and mammalian cultured cells. Further, we are trying to identify causative genes for hereditary human myopathies using fly genetics.
We are also working on analyzing the mechanism and function of autophagy-mediated phenomena through a unique approach using Drosophila.
Selected publications
- 1.Homma, Y., Kinoshita, R., Kuchitsu, Y., Wawro, P. S., Marubashi, S., Oguchi, M., Ishida, M., Fujita, N., Fukuda, M. Comprehensive knockout analysis of the Rab family GTPases in epithelial cells.J Cell Biol (2019) 218, 2035-2050.
- 2.Kuchitsu, Y., Homma, Y., Fujita, N.#, and Fukuda, M.# Rab7 knockout unveiled regulated autolysosome maturation induced by glutamine starvation J Cell Sci (2018) 131, jcs215442 (# co-corresponding authors)
- 3.Fujita, N.#, Huang, W., Lin, T.H., Groulx, J.F., Jean, S., Nguyen, J., Kuchitsu, Y., Koyama-Honda, I., Mizushima, N., Fukuda, M., and Kiger, A.A.# Genetic screen in Drosophila muscle identifies autophagy-mediated T-tubule remodeling and a Rab2 role in autophagy. Elife (2017) 6. e23367 (# co-corresponding authors)
- 4.Fujita, N.*, Morita, E.*, Itoh, T., Tanaka, A., Nakaoka, M., Osada, Y., Umemoto, T., Saitoh, T., Nakatogawa, H., Kobayashi, S., Haraguchi, T., Guan, J.L., Iwai, K., Tokunaga, F., Saito, K., Ishibashi, K., Akira, S., Fukuda, M., Noda, T., and Yoshimori, T. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol (2013) 203, 115-128. (* co-first authors)
- 5.Hamasaki, M., Furuta, N., Matsuda, A., Nezu, A., Yamamoto, A., Fujita, N., Oomori, H., Noda, T., Haraguchi, T., Hiraoka, Y., Amano, A., and Yoshimori, T. Autophagosomes form at ER-mitochondria contact sites. Nature (2013) 495, 389-393.
- 6.Fujita, N., Saitoh, T., Kageyama, S., Akira, S., Noda, T., and Yoshimori, T. Differential involvement of Atg16L1 in Crohn disease and canonical autophagy: analysis of the organization of the Atg16L1 complex in fibroblasts. J Biol Chem (2009) 284, 32602-32609.
- 7.Hayashi-Nishino, M., Fujita, N., Noda, T., Yamaguchi, A., Yoshimori, T., and Yamamoto, A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol (2009) 11, 1433-1437.
- 8.Saitoh, T.*, Fujita, N.*, Hayashi, T., Takahara, K., Satoh, T., Lee, H., Matsunaga, K., Kageyama, S., Omori, H., Noda, T., Yamamoto, N., Kawai, T., Ishii, K., Takeuchi, O., Yoshimori, T., and Akira, S. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response.
Proc Natl Acad Sci U S A (2009) 106, 20842-20846. (* co-first authors) - 9.Saitoh, T. *, Fujita, N.*, Jang, M.H., Uematsu, S., Yang, B.G., Satoh, T., Omori, H., Noda, T., Yamamoto, N., Komatsu, M., Tanaka, K., Kawai, T., Tsujimura, T., Takeuchi, O., Yoshimori, T., and Akira, S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature (2008) 456, 264-268. (* co-first authors)
- 10.Fujita, N., Hayashi-Nishino, M., Fukumoto, H., Omori, H., Yamamoto, A., Noda, T., and Yoshimori, T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell (2008) 19, 4651-4659.
- 11.Fujita, N., Itoh, T., Omori, H., Fukuda, M., Noda, T., and Yoshimori, T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell (2008) 19, 2092-2100.
Contact
Associate Professor, Naonobu Fujita
Room 210, S2 building, Suzukakedai campus
e-mail:nafujita@bio.titech.ac.jp
*Find more about the lab and the latest activities at Fujita Lab.![]() (Japanese)
(Japanese)
*May 1, 2025:Some of the content has been updated with the latest information.