Life Science and Technology News
【Labs spotlight】 Akio Kitao Laboratory
Functional simulation of bio-nanomachine
The Department has a variety of laboratories for Life Science and Technology, in which cutting-edge innovative research is being undertaken not only in basic science and engineering but also in the areas of medicine, pharmacy, agriculture, and multidisciplinary sciences.
This "Spotlight" series features a laboratory from the Department and introduces you to the laboratory's research projects and outcomes. This time we focus on Akio Kitao Laboratory.

Areas of Supervision
Primary/Life Science and Technology
Professor Akio Kitao![]()
| Degree | PhD 1994, Kyoto University |
|---|---|
| Areas of Research | Computational biology, Biophysics, Computational chemistry |
| Keywords | Protein dynamics, Molecular dynamics, Protein docking |
| Website | Akio Kitao Lab. |
Research interest
Observing Biological Phenomena by Computer
Biological phenomena can be observed by various experimental methods; however, to investigate such phenomena at molecular level, observations at atomic resolution are required. We simulate biological events at atomic resolution, which cannot be observed by experiments, by State-of-the-art computer simulations, and investigate functional mechanisms of biomolecules.
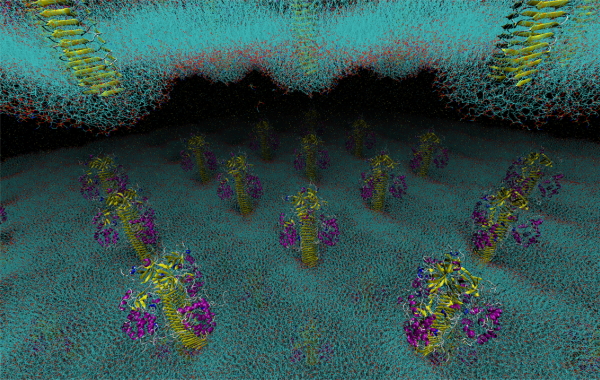
Functional simulation of bio-nanomachine
We investigate functional mechanism of biological nano-machines which consist of biomolecules. We simulate the behavior of biological system including nano-machines and solvent molecules in silico in atomic detail by using molecular dynamics (MD) simulation.
We show an example of bacterial flagellar motor. The bacterial flagellum is a molecular machine for the locomotion of bacteria. The flagellar motor can convert the motive force of protons and other ions to molecular motor rotations. We investigated the multi-ion transfer mechanism of the flagellar motor stator MotA/B in Escherichia coli, which primarily utilizes protons. We also showed that the protonation/deprotonation cycle induces a ratchet motion in the stator, which can couple to the motor rotation. Ion selectivity and the ratchet motion are key to understanding the mechanism of the flagellar motor.
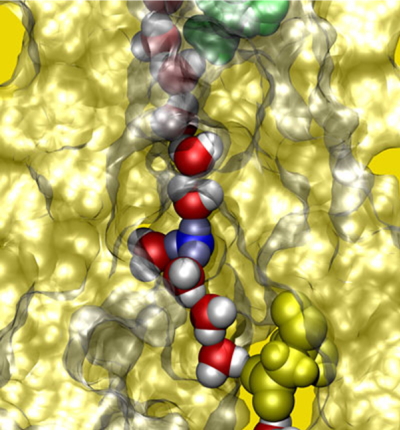
Development of more efficient and accurate simulation methods
We have been developing new molecular simulation methods by which system containing multiple biopolymers can be simulated efficiently. Also, we have been developing the methods to analyze simulation results and extract functionally important conformational motion in proteins.
For example, we have developed Parallel Cascade Selection Molecular Dynamics (PaCS-MD), which is an efficient conformational sampling method for biomolecular systems. In PaCS-MD, cycles of selection of initial structures for multiple independent MD simulations and conformational sampling by independent MD simulations are repeated until the convergence of the sampling. The selection of snapshots enhances conformational changes by increasing the probability of rare event occurrence toward the target. PaCS-MD enables us to simulate large conformational change of protein, and dissociation/association dynamics of protein complexes.
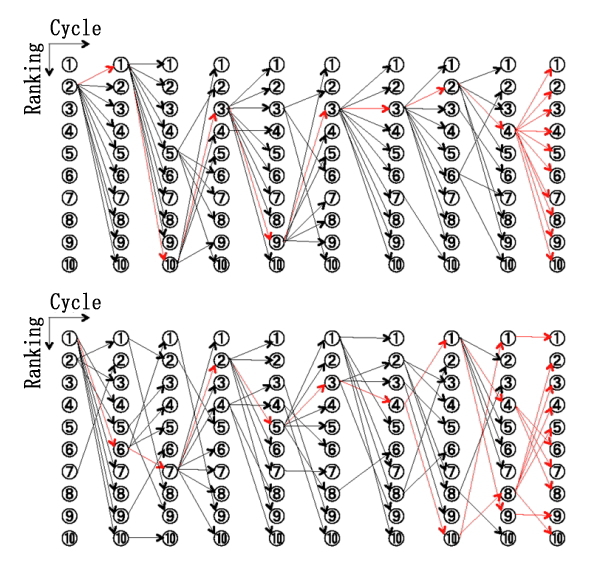
Dissociation and association dynamics of biomolecules
Association and dissociation of biomolecules are essential biological processes; however, they have not been well characterized yet. We have been investigating association and dissociation mechanism of biomolecules by molecular simulation. This type of research is also important for drug discovery. PaCS-MD enables us to simulation biomolecular association and dissociation processes and to estimate binding affinity and kinetics.
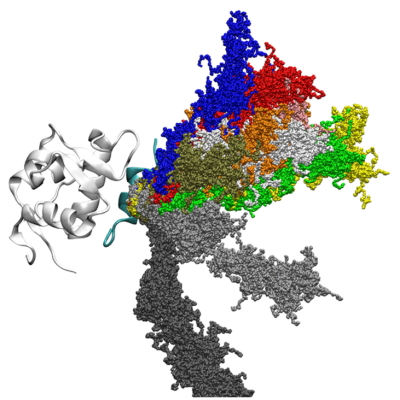
Future of molecular simulation
If advancements of computers and simulation methods continue, we would expect that molecular simulation can conduct quantitative predictions before performing experiments in the future.
Selected publications
Publication in English
http://www.researcherid.com/rid/B-8371-2008![]()
Contact
Professor Akio Kitao
Room 201C, Midorigaoka Building 6, Ookayama campus
Email akitao@bio.titech.ac.jp
*Find more about the lab and the latest activities at Kitao Lab.![]()
*October 16, 2025:Some of the content has been updated with the latest information.