Life Science and Technology News
Highly sensitive Raman probe detects enzyme expression in heterogeneous tissues
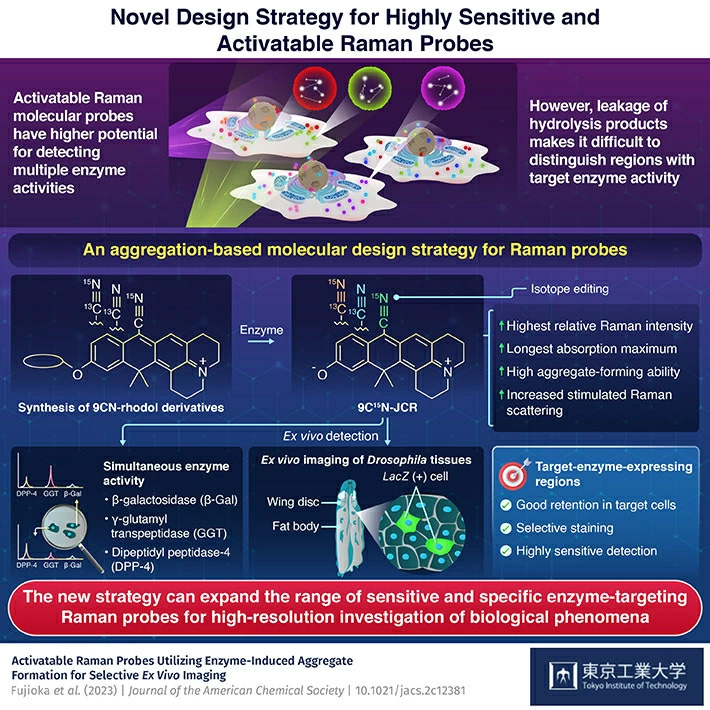
Raman imaging offers a greater potential for detecting multiple enzyme activities than fluorescence imaging, demonstrate Tokyo Tech researchers by developing 9CN-rhodol-based activatable Raman probes using a novel mechanism for Raman signal activation. The strategy allows a synthesis of highly activatable Raman probes with high aggregation and multiplexing ability, making it a promising tool for extending the range of Raman probes for the detection of multiple enzyme activities in heterogeneous biological tissues.
The involvement of enzymes in a wide range of biological activities makes them ideal biomarkers for the detection of diseases. In fact, cancer-specific diagnostic technologies use fluorescence imaging for detecting upregulated cancer-associated enzymes in the affected cells. Moreover, since tumor tissues are heterogenous, detecting multiple enzyme activities simultaneously could allow precise cancer visualization and diagnosis. However, the inability to detect multiple enzyme activities can potentially limit the application of fluorescence imaging in heterogeneous tumor tissues and other complex biological phenomena.
As an alternative, the narrower spectral width of Raman spectral imaging offers hope for multiplexing biological imaging with molecular probes. Over the years, several functional and activatable Raman probes (dyes) for detecting bioanalytes have been developed, but those for detecting enzyme activities have been limited. Further, the previous design strategies have failed to control the diffusion of enzyme-generated hydrolysis product of these probes, making it difficult to distinguish regions with target enzyme activity in tissues.
Against this backdrop, a team of researchers from Japan led by Professor Mako Kamiya and Assistant Professor Hiroyoshi Fujioka of Tokyo Institute of Technology (Tokyo Tech) recently reported, taking inspiration from aggregation-based fluorescent probes, a new molecular design strategy for the synthesis of activatable Raman probes based on 9CN-rhodol. Their study was published in the Journal of the American Chemical Society![]() .
.
Explaining the choice of rhodol derivatives for molecular scaffold, Prof. Kamiya says, "Hiroyoshi found that rhodol derivatives with a nitrile group at the ninth position (9CN-rhodols) and a net charge of zero exhibit a single sharp Raman peak and have higher aggregation ability in aqueous solution than 9CN-pyronins with positive charge. Accordingly, we used 9CN-rhodol scaffold dyes to create Raman probes that can exhibit high sensitivity and undergo red shift in molecular absorption towards a region of resonant Raman effect and form aggregation upon interacting with the target enzymes ."
Accordingly, the team first synthesized 9CN-rhodol derivatives and selected two derivatives, 9CN-JR and 9CN-JCR, as candidates for designing the activatable Raman probes. They then tested the enzyme detection performance of both probes in live cells using a dual-color stimulated Raman scattering (SRS) imaging technique. Of the two, 9CN-JCR emerged as the better and brighter probe for multiplexing.
Next, the team isotope-labeled the nitril group of 9CN-JCR scaffold with Carbon-13 (13C) and Nitrogen-15 (15N) and then created two new isotope edited 9CN-JCR probes for γ-glutamyl transpeptidase and dipeptidyl peptidase-4 enzymes. The new set of 9CN-JCR-based probes was then able to simultaneously detect the activities of all these enzymes in the live cell culture.
Moreover, the probes allowed ex vivo imaging of distinct cell regions expressing target enzyme activity in Drosophila wing disk and fat body. The high spatial selectivity and sensitivity exhibited by the 9CN-JCR probes was attributed to the electronic pre-resonance effect of the scaffold dye and aggregate formation of the hydrolysis products formed by probe-target cell interaction.
The rhodol-based probes could aggregate upon reaction with the enzymes, improving their intracellular retention, and increased the SRS signal intensity during enzyme detection.
In summary, the facile strategy demonstrated in this study can facilitate the development of highly specific activatable Raman probes for the simultaneous detection of multiple enzyme activities. "Our aggregation-based molecular design strategy for Raman probes will offer substantial advantages for applications involving the investigation of enzyme activity associated with diseases and essential biological activities," concludes Prof. Kamiya.
This project was conducted in collaboration with Prof. Yasuyuki Ozeki at The University of Tokyo.
- Reference
| Authors : | Hiroyoshi Fujioka 1,2, Minoru Kawatani 2,3, Spencer John Spratt 4, Ayumi Komazawa 1, Yoshihiro Misawa 2,3, Jingwen Shou 4, Takaha Mizuguchi 4, Hina Kosakamoto 5, Ryosuke Kojima 3, Yasuteru Urano 1,3, Fumiaki Obata 5,6, Yasuyuki Ozeki 4, and Mako Kamiya 2,3,7 |
|---|---|
| Title : | Activatable Raman Probes Utilizing Enzyme-Induced Aggregate Formation for Selective Ex Vivo Imaging |
| Journal : | Journal of the American Chemical Society |
| DOI : | |
| Affiliations : | 1 Graduate School of Pharmaceutical Sciences, The University of Tokyo 2 Department of Life Science and Technology, Tokyo Institute of Technology 3 Graduate School of Medicine, The University of Tokyo 4 Department of Electrical Engineering and Information Systems, Graduate School of Engineering, The University of Tokyo 5 RIKEN Center of Biosystems Dynamics Research 6 Graduate School of Biostudies, Kyoto University 7 Living Systems Materialogy (LiSM) Research Group, International Research Frontiers Initiative (IRFI), Tokyo Institute of Technology |
- Kamiya Laboratory
- Mako Kamiya | Researcher Finder - Tokyo Tech STAR Search
- Department of Life Science and Technology, School of Life Science and Technology
- International Research Frontiers Initiative (IRFI), Tokyo Institute of Technology
- Graduate School of Pharmaceutical Sciences, Faculty of Pharmaceutical Sciences, The University of Tokyo
- Graduate School of Medicine and Faculty of Medicine, The University of Tokyo
- Department of Electrical Engineering and Information Systems, Graduate School of Engineering, The University of Tokyo
- RIKEN Center for Biosystems Dynamics Research (RIKEN BDR)
- Latest Research News
School of Life Science and Technology
—Unravel the Complex and Diverse Phenomena of Life—
Information on School of Life Science and Technology inaugurated in April 2016
Further Information
Professor Mako Kamiya
International Research Frontiers Initiative (IRFI), Tokyo Institute of Technology
E-mail : kamiya.m.ad@m.titech.ac.jp
Tel : +81-45-924-5786