Life Science and Technology News
Not so selfish after all ― Key role of transposable elements in mammalian evolution
A scientist at Tokyo Institute of Technology (Tokyo Tech) has revealed a key role for "selfish" transposable elements in the evolution of the mammary gland, a defining feature of all mammals.
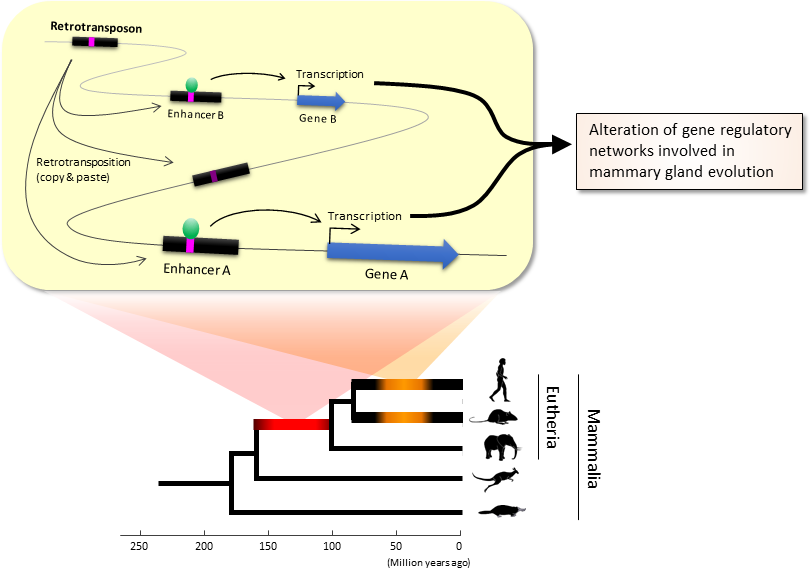
- Figure 1.
- Retrotransposons have been co-opted to act as enhancers in the mammalian genome.
- Transposable elements known as retrotransposons duplicate themselves via a copy-and-paste mechanism. Tens of thousands of these sequences were found to contain binding sites for proteins that act as master regulators of mammary gland development.
The human genome contains 4.5 million copies of transposable elements (TEs), so-called selfish DNA sequences capable of moving around the genome through cut-and-paste or copy-and-paste mechanisms. Accounting for 30-50% of all of the DNA in the average mammalian genome, these TEs have conventionally been viewed as genetic freeloaders, hitchhiking along in the genome without providing any benefit to the host organism. More recently, however, scientists have begun to uncover cases in which TE sequences have been co-opted by the host to provide a useful function, such as encoding part of a host protein. In a new study published in the journal Nucleic Acids Research, Professor Hidenori Nishihara has undertaken one of the most comprehensive analyses of TE sequence co-option to date, uncovering tens of thousands of potentially co-opted TE sequences and suggesting that they have played a key role in mammalian evolution.
"I was specifically interested in the potential influence of TE sequences on the evolution of the mammary gland," notes Dr. Nishihara, "an organ that is responsible for producing milk and is, as the name suggests, a key distinguishing feature of mammals." To identify potentially co-opted TE sequences, Dr. Nishihara used four proteins-ERα, FoxA1, GATA3, and AP2γ-that bind to DNA to regulate the production of proteins involved in mammary gland development. Dr. Nishihara then located all of the DNA sequences in the genome to which these proteins bind. Surprisingly, 20-30% of all of the binding sites across the genome were located in TEs, with as many as 38,500 TEs containing at least one binding site. The majority of these were in a copy-and-paste type of TE known as a retrotransposon, which duplicates itself, leaving a new copy in a new location.
The TE-derived binding site sequences were more conserved across species than expected, indicating that they are being preserved by evolution because they serve some important function. Dr. Nishihara believes that these TE sequences have been co-opted to serve as enhancers, DNA elements that increase the transcription of nearby genes (Fig. 1). By binding to one of the four master regulators of mammary gland development, these enhancers ultimately increase the production of proteins involved in mammary gland development.
Dr. Nishihara then investigated when in mammalian evolution these TE sequences were acquired and found two distinct phases of acquisition: roughly 60-70% were acquired in the ancestor of all placental mammals (Eutheria), while 10-20% could be traced back to the ancestor of New World monkeys (Simiiformes) (Fig. 2, left). In addition, there appeared to be another wave of acquisition of ERα binding sites in the ancestor of mice and rats (Muridae) (Fig. 2, right). Thus, by providing a vast number of potential regulatory element binding sites throughout the genome, TEs may have had a substantial impact on the emergence of the mammary gland and its evolution within mammals.
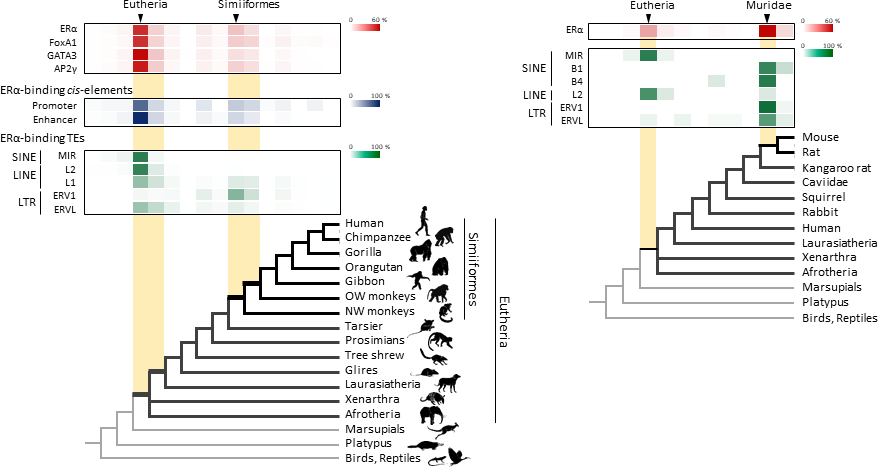
- Figure 2.
- Transposable element-derived binding sites were acquired during distinct phases in mammalian evolution.
- Left: Among the TE-derived binding sites identified, 60-70% were acquired in the ancestor of placental mammals (Eutheria), while 10-20% were acquired in the ancestor of New World monkeys (Simiiformes). Right: Many ERα binding sites were also acquired in the ancestor of mice and rats (Muridae).
Dr. Nishihara's study sheds light on the deep involvement of TEs in the evolution of mammary gland regulatory elements. However, it remains unclear how common this mode of TE-mediated regulatory network evolution is. Dr. Nishihara, at least, believes that the mammary gland is not unique in this respect. He notes that, "in addition to mammary glands, mammals share many features, such as the neocortex, closed secondary palate, and hair. I expect future research to uncover many additional kinds of TEs that have been similarly involved in the evolution of these features in mammals."
- Reference
| Authors : | Hidenori Nishihara |
|---|---|
| Title of original paper : | Retrotransposons spread potential cis-regulatory elements during mammary gland evolution |
| Journal : | Nucleic Acids Research |
| DOI : | 10.1093/nar/gkz1003 |
| Affiliations : | Department of Life Science and Technology, Tokyo Institute of Technology |
- Past genetic polymorphisms contributed to African cichlid diversity
- KIMURA LAB (Japanese)
- Researcher Profile | Tokyo Tech STAR Search - Hidenori Nishihara
- Department of Life Science and Technology, School of Life Science and Technology
- Latest Research News
School of Life Science and Technology
—Unravel the Complex and Diverse Phenomena of Life—
Information on School of Life Science and Technology inaugurated in April 2016
Further Information
Assistant Professor Hidenori Nishihara School of Life Science and Technology, Tokyo Institute of Technology
E-mail : hnishiha@bio.titech.ac.jp
Tel +81-45-924-5742