Life Science and Technology News
A rapid, easy-to-use DNA amplification method at 37℃
Study could advance molecular diagnosis and robotics
Scientists in Japan have developed a way of amplifying DNA on a scale suitable for use in the emerging fields of DNA-based computing and molecular robotics. By enabling highly sensitive nucleic acid detection, their method could improve disease diagnostics and accelerate the development of biosensors, for example, for food and environmental applications.
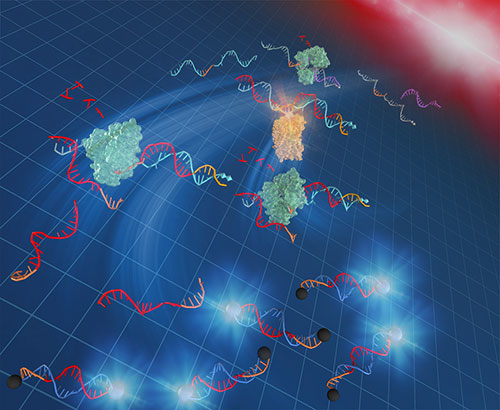
Figure 1. Schematic illustration of L-TEAM and targeted hybridization
A DNA strand (purple) primes exponential amplification of DNA (red) as signals for directing light emission of DNA nanodeveices.
Researchers from Tokyo Institute of Technology (Tokyo Tech), Abbott Japan Co., Ltd, and the University of Electro-Communications, Japan, report a way to achieve million-fold DNA amplification and targeted hybridization1 that works at body temperature (37℃/98.6°F).
The method, named L-TEAM (Low-TEmperature AMplification), is the result of more than five years of research and offers several advantages over traditional PCR2, the dominant technique used to amplify DNA segments of interest.
With its easy-to-use, ‘one-pot’ design, L-TEAM avoids the need for heating and cooling steps and specialized equipment usually associated with PCR. That means it is an efficient, inexpensive method that can importantly prevent protein denaturation3, thereby opening a new route to real-time analysis of living cells.
In their study published in Organic & Biomolecular Chemistry, the researchers introduced synthetic molecules called locked nucleic acids (LNAs) into the DNA strands, as these molecules are known to help achieve greater stability during hybridization.
The addition of LNA led to an unexpected, but beneficial, outcome. The team observed a reduced level of "leak" amplification, a type of non-specific amplification that has long been an issue in DNA amplification studies as it can lead to an error in disease diagnosis, that is, a false positive.
"We were surprised to discover the novel effect of LNA in overcoming the common leak problem in DNA amplification reactions," says Ken Komiya, assistant professor at Tokyo Tech's School of Computing. "We plan to investigate the mechanisms behind leak amplification in detail and further improve the sensitivity and speed of L-TEAM."
In the near future, the method could be used to detect short nucleic acids such as microRNA for medical diagnostics. In particular, it could facilitate point-of-care testing and early disease detection. MicroRNAs are now increasingly recognized as promising biomarkers for cancer detection and may hold the key to uncovering many other aspects of human health and environmental science.
In addition, Komiya explains that L-TEAM paves the way to practical use of DNA computing and DNA-controlled molecular robotics. "The original motivation behind this work was the construction of a novel amplified module that is essential to build advanced molecular systems," he says. "Such systems could provide insights into the operational principle behind living things."
The annealing or binding of single-stranded DNA to a complementary strand.
Polymerase chain reaction, a widely used technique to make multiple copies of a segment of DNA.
The breakdown of bonds that hold a protein in its native structure.
Reference
| Authors : | K. Komiyaa, M. Komorib, C. Nodaa, S. Kobayashic, T. Yoshimurab and M. Yamamuraa |
|---|---|
| Title of original paper : | Leak-free million-fold DNA amplification with locked nucleic acid and targeted hybridization in one pot |
| Journal : | Organic and Biomolecular Chemistry, 21 June 2019, issue 23, pp 5708-5713 (Featured as the outside front cover). Published online 9 April 2019. |
| DOI : | |
| Affiliations : |
|
- Yamamura Lab.
- Komiya group at Yamamura-Lab
- Researcher Profile | Tokyo Tech STAR Search - Ken Komiya
- Researcher Profile | Tokyo Tech STAR Search - Masayuki Yamamura
- Department of Computer Science, School of Computing
- Molecular Robotics
- The University of Electro-Communications
- Latest Research News
Further Information
Assistant Professor Ken Komiya
School of Computing
Tokyo Institute of Technology
E-mail : komiya@c.titech.ac.jp
Tel +81-3-5734-2975