Life Science and Technology News
When less is more: A promising approach for low-cell-number epigenomic profiling
Scientists at Kyushu University and Tokyo Tech in Japan have developed a technique that enables analysis of DNA-protein interactions using very small numbers of cells, ranging from 100 to 1,000. Their method could capture previously unexamined epigenomic information, facilitate biomarker discovery and open new avenues for precision medicine.
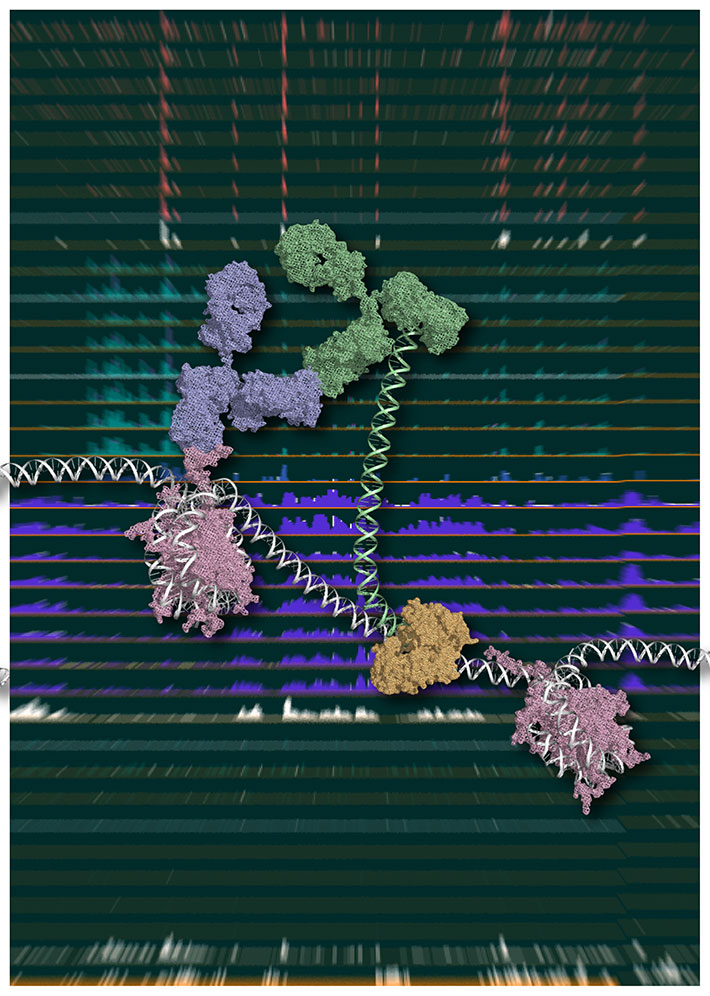
Figure 1. Chromatin Integration labeling (ChIL)
Schematic model of ChIL reaction. The oligonucleotide conjugated with the secondary antibody (green), which binds to the primary antibody (blue) recognizing a specific medication of histone (purple), is integrated into proximal genomic DNA by Tn5 transposase (brown). An image courtesy of Yuko Sato (Tokyo Institute of Technology).
A collaborative study by researchers at Kyushu University, Tokyo Institute of Technology, Waseda University, the University of Tokyo and Osaka University has led to the development of a unique approach to epigenomic profiling1 that involves working with much fewer cells than in existing methods.
The technique, called Chromatin Integration Labeling sequencing (ChIL-seq), could open up new opportunities for scientists to study rare cell types and other cell samples in short supply. ChIL-seq requires only a fraction of starting cellular material. In a series of experiments to evaluate ChIL-seq performance, the researchers successfully demonstrated detection of histone modifications2 and DNA-binding factors using just 100 to 1,000 cells.
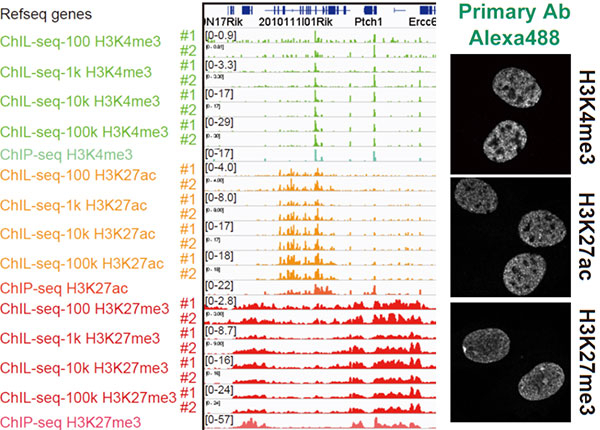
Figure 2. Chromatin Integration labeling sequencing
Histone modifications or transcription factors are labeled with ChIL probe in cell nuclei (right), and the genomic sequence proximal to the probe is amplified for large scale sequencing analysis (left). The profiles observed in 100 cells are similar to those using more than 1,000 cells.
For the past decade, chromatin immunoprecipitation sequencing (ChIP-seq) has been the dominant technique for analyzing epigenomic data and identifying important binding sites of DNA-associated proteins. However, one limitation has been that ChIP-seq requires at least 10,000 or typically millions of cells to start off with, mainly due to the fact that samples tend to be lost during two key steps: chromatin preparation3 and immunoprecipitation4.
The research team, co-led by Hiroshi Kimura of the Institute of Innovative Research, Tokyo Institute of Technology, and Yasuyuki Ohkawa of Medical Institute of Bioregulation, Kyushu University, overcame the problem of sample loss by replacing the two steps mentioned above with immunostaining, a non-destructive method suitable for analyzing tissue specimens.
Using ChIL-seq, the team also detected genomic regions associated with histone marks at the single-cell level -- an achievement that brings biologists closer to the long-held goal of establishing single-cell profiling.
ChIL-seq can “zoom in” on genomic sequences near target molecules before cellular breakdown, and this is particularly useful for studying adherent cells, i.e., whole cells that remain attached to culture plates and after immunofluorescence.
Many different types of epigenome profiling methods are now being developed around the world. Each has its advantages and limitations. The researchers point out that ChIL-seq, too, still needs refining. In its current form, for example, it has low sensitivity to heterochromatin regions, and it can take 3-4 days to complete the procedure.
Overall, they are confident that ChIL-seq holds promise due to its precision, which makes it suitable for single-cell applications, and its flexibility, meaning that in future, it could be combined with other powerful sequencing techniques.
Comments from researchers

Prof. Yasuyuki Ohkawa of Kyushu University (left) and Prof. Hiroshi Kimura of Tokyo Tech
We as epigenome researchers had been in great need of this technology. Though we established a new conception earlier, it took us more than five years to eventually become a practical technique. We strongly hope this technology will help great leap in life science, including the regenerative medicine by stem cells and elucidation of mechanism of cancers.
Analysis of DNA-protein interactions that can provide insights into disease states and therapeutic targets.
Post-translational modifications that regulate gene expression.
A process involving the extraction of information-rich chromatin (a complex of DNA and proteins) through the breakdown of cells.
Purification of target proteins based on the antigen-antibody interaction.
Reference
| Authors : | Akihito Harada1, Kazumitsu Maehara1, Tetsuya Handa2, Yasuhiro Arimura3,4, Jumpei Nogami1, Yoko Hayashi-Takanaka5,6, Katsuhiko Shirahige4, Hitoshi Kurumizaka3,4 Hiroshi Kimura2,5 Yasuyuki Ohkawa1 |
|---|---|
| Title of original paper : | A chromatin integration labelling method enables epigenomic profiling with lower input |
| Journal : | Nature Cell Biology (2018) |
| DOI : | 10.1038/s41556-018-0248-3 |
| Affiliations : |
1Division of Transcriptomics, Medical Institute of Bioregulation, Kyushu University 2Cell Biology Center, Institute of Innovative Research, Tokyo Institute of Technology 3Laboratory of Structural Biology, Graduate School of Advanced Science and Engi-neering, Research Institute for Science and Engineering, and Institute for Medical-oriented Structural Biology, Waseda University 4Institute for Quantitative Biosciences, the University of Tokyo 5Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology 6Graduate School of Frontier Biosciences, Osaka University |
| Corresponding author's email : | hkimura@bio.titech.ac.jp,yohkawa@bioreg.kyushu-u.ac.jp |
- Catching histones by the tail: a new probe to track histone modifications in living cells | Life Science and Technology News
- Professor Hiroshi Kimura Receives 2015 Robert Feulgen Prize | Tokyo Tech News
- KIMURA LAB (japanese)
- Researcher Profile | Tokyo Tech STAR Search - Hiroshi Kimura
- Researcher Profile | Tokyo Tech STAR Search - Tetsuya Handa
- Cell Biology Center Institute of Innovative Research Tokyo Institute of Technology
- Institute of Innovative Research (IIR)
- Department of Life Science and Technology, School of Life Science and Technology
- Medical Institute of Bioregulation Kyushu University
- Japan Science and Technology Agency
- IQB, The University of Tokyo
- Latest Research News
Contact
Public Relations Section, Tokyo Institute of Technology
E-mail : media@jim.titech.ac.jp
Tel +81-3-5734-2975