Life Science and Technology News
Target of rapamycin: linking cytosolic and chloroplast ribosome biogenesis in plants
Scientists at Tokyo Institute of Technology (Tokyo Tech) have found that eukaryotic and bacterial growth regulation systems of independent origins are connected to the control of chloroplast rRNA transcription in a primitive red alga.
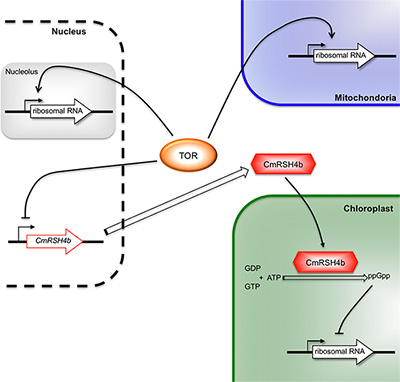
Figure 1. The regulatory mechanism for biogenesis
A linked regulatory mechanism for biogenesis of the three ribosomes by TOR.
Chloroplasts are crucial specialized structures within a cell that generate energy and produce storage- and cell metabolism-related molecules that underpin cell growth. The reactions that take place in chloroplasts require the formation of proteins by the chloroplast ribosome (site of protein synthesis in the organelle); however, how this is regulated in plant cells is still largely unknown. The conversion of ribosomal DNA to ribosomal RNA (rRNA), i.e., transcription, is thought to be a rate-limiting step of biogenesis (the slowest step that determines the speed of the ribosome biogenesis), and hence it is crucial to clearly understand rRNA transcription.
Previous studies have revealed that inhibiting TOR (target of rapamycin), a highly conserved protein kinase, represses nuclear rRNA transcription in many eukaryotes, and hence Associate Professor Sousuke Imamura and colleagues hypothesized that TOR regulates rRNA transcription in chloroplasts and mitochondria. They found that TOR modulates the transcription of chloroplast rRNA, as well as that of nuclear and mitochondrial rRNA, in a model unicellular red alga, Cyanidioschyzon merolae. The result indicates that TOR is involved in a mechanism that regulates biogenesis in all three ribosomes.
But how does TOR regulate chloroplast rRNA transcription in a cell? This group of researchers tried to answer this question by analyzing the effects of TOR inhibition. They found that this inhibition increased the expression of a nuclear-encoded chloroplast gene, CmRSH4b, which encodes a homolog of the signal molecule guanosine 3′-diphosphate 5′-diphosphate (ppGpp) synthetases that modulates rRNA synthesis in bacteria. Further genetic and biochemical analyses demonstrated that the formation of CmRSH4b-dependent ppGpp in chloroplasts is an important regulator of chloroplast rRNA transcription.
Thus, the results of this study lay the foundation for future research in chloroplast ribosome biogenesis. Understanding the coordination between the three interconnected ribosome systems will allow for further insights into the regulation of growth of algal and complex plant cells.
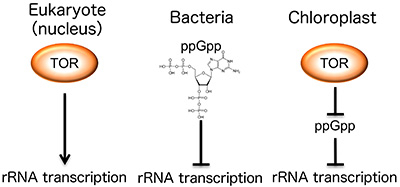
Figure 2. The two systems combine to regulate chloroplast rRNA transcription
Eukaryotic and bacterial growth regulation systems of independent origins work in unison to control chloroplast rRNA transcription.
Reference
| Authors : | Sousuke Imamura1, Yuhta Nomura2, Tokiaki Takemura1, Imran Pancha1, Keiko Taki1, Kazuki Toguchi1, Yuzuru Tozawa2, Kan Tanaka1 |
|---|---|
| Title of original paper : | The checkpoint kinase TOR (target of rapamycin) regulates expression of a nuclear-encoded chloroplast RelA-SpoT homolog (RSH) and modulates chloroplast ribosomal RNA synthesis in a unicellular red alga |
| Journal : | The Plant Journal |
| DOI : | 10.1111/tpj.13859 |
| Affiliations : |
1Laboratory for Chemistry and Life Science, Institute of Innovative Research, Tokyo Institute of Technology 2Graduate School of Science and Engineering, Saitama University |
- Algal residue - an alternative carbon resource for pharmaceuticals and polyesters|Tokyo Tech News
- Shedding light on phototactic behavior of green algae|Tokyo Tech News
- Abscisic Acid Helps Red Algae Tolerate Salt Stress by Controlling Cell Cycle Initiation|Tokyo Tech News
- TANAKA-IMAMURA Lab,TTECH
- Researcher Profile | Tokyo Tech STAR Search - Kan Tanaka
- Researcher Profile | Tokyo Tech STAR Search - Sousuke Imamura
- Laboratory for Chemistry and Life Science
- Institute of Innovative Research(IIR)
- Department of Life Science and Technology, School of Life Science and Technology
- Latest Research News
Further Information
Associate Professor Sousuke Imamura
Laboratory for Chemistry and Life Science,
Institute of Innovative Research, Tokyo Institute of Technology
Email simamura@res.titech.ac.jp
Tel +81- 45-924-5859