Life Science and Technology News
【Labs spotlight】 Tanaka and Imamura Laboratory
From Understanding to Design of Cell Systems
The Department has a variety of laboratories for Life Science and Technology, in which cutting-edge innovative research is being undertaken not only in basic science and engineering but also in the areas of medicine, pharmacy, agriculture, and multidisciplinary sciences.
This "Spotlight" series features a laboratory from the Department and introduces you to the laboratory's research projects and outcomes. This time we focus on Tanaka and Imamura Laboratory.

Areas of Supervision
Primary/Life Science and Technology
Secondary/Human Centered Science and Biomedical Engineering
Professor Kan Tanaka![]()

Life Science and Technology
Associate professor Sousuke Imamura![]()
*February 28, 2021:Associate Professor Imamura retired from Tokyo Tech.
| Degree | Professor Kan Tanaka : PhD 1990, The University of Tokyo Associate professor Sousuke Imamura : PhD 2005, Tokyo University of Agriculture and Technology |
|---|---|
| Areas of Research | Evolutional Cell Biology, Microbiology, Molecular Genetics, Plant Molecular Biology, Genetic engineering. |
| Keywords | Algal Biomass, Chloroplast, Cyanidioschyzon merolae, Cyanobacteria, Escherichia coli, Eukaryotic Algae, Metabolism, Photosynthesis, Organelle Signal, Ribosome, Sigma Factor, Signal Transduction, Symbiosis, Transcription, Organelle Communication, Ribosome RNA, Transcriptional Regulation |
| Website | Tanaka and Imamura Laboratory |
Research interest
Professor Kan Tanaka
A) Eukaryotic cell has emerged through the symbiotic association of prokaryotic cells. Plant cell has also evolved as the result of the cyanobacterial endosymbiosis. Providing that endosymbiotic association has established the eukaryotic framework, we are interested in the understanding of various aspects of eukaryotic cell biology, such as cell cycle and environmental acclimation, based on the inter-organelle communications.
B) Eukaryotic cell was constructed based on the association of prokaryotic cells, and thus prokaryotic cells are the real basal unit of every living cell. We are interested in the basic architect of bacterial cells, focusing on metabolism, ribosome, genome and their interrelationship.
C) Oxygenic photosynthesis is the fundamental process for the earth biosystem, which was evolved by ancient cyanobacteria and descended to plant chloroplasts. Since the photosynthetic machineries are labile to environmental changes, photosynthetic organisms have established the acclimation system to protect the system from various damages. We are studying the conserved core part of the protection mechanism from the aspect of the environmental sensing and the signal transduction.
D) Photosynthetic microorganisms are able to accumulate useful biomass as oils under stressful conditions. We are working to understand the underlying mechanism for the production to establish the basis for the breeding of industrially useful organisms.
Associate professor Sousuke Imamura
Nitrogen is an important nutrient for all organisms, as the nutrient is composed of the major biological molecules such as amino acids and nucleic acids. Therefore, research on control of nitrogen metabolism has been extensively studied in eukaryotes, and revealed many important regulatory factors and signal transductions that are involved in nitrogen regulation. However, the related information in plant cells are very poor.
We are mainly striving to clarify the fundamental molecular mechanisms of responses to nitrogen deficiency in Cyanidioschyzon merolae. C. merolae is a unicellular red alga, with each cell containing only one mitochondrion, one chloroplast, and one nucleus. The complete genome sequences of these three organelles were determined, and their simple, minimally redundant gene content was revealed. Because of the elucidation of these biological characteristics and the establishment of various tools for analysis of C. merolae, this alga is thought to be a good model organism to understand various molecular functions of photosynthetic eukaryotes.
So far, we have revealed enzymes and pathways that contribute to nitrogen assimilation [Imamura et al (2010) Plant Cell Physiol]. Furthermore, we have identified and characterized a transcription factor, named MYB1, which is indispensable for gene expression of nitrogen assimilation [Imamura et al (2009) PNAS]. However, those points are only part of nitrogen regulation in plant cells [Imamura et al (2015) Plant Mol Biol]. For example, how plant cells sense nitrogen levels and quality, and how they transmit the information to organelles are entirely unknown. So, we are trying to solve the problem using the alga.
Microalgae highly accumulate good feedstock for biofuel production, such as triacylglycerol (TAG) and starch, in response to nitrogen deficient conditions. Although the algal biomass are considered as a renewable energy resources, improvement of productivity of biomass production is required to establish a commercial production of biofuel [Imamura et al (2016) Plant Signal Behav]. Thus, we are also doing research that applies the knowledge of fundamental nitrogen regulation to efficient production of algal biomass using genetic engineering approach.
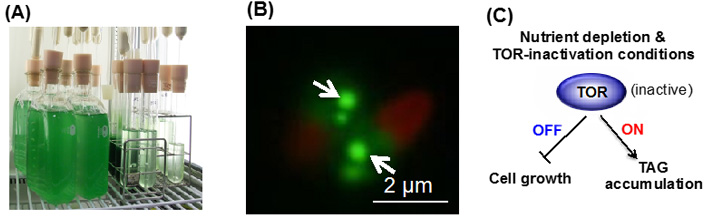
Algal culture (A)
Accumulation of TAG (arrows) by TOR-inactivation (B)
and its regulatory model (C)
Research result
Selected publications
Professor Kan Tanaka
- 1.Zeenat B. Noordally, Kenyu Ishii, Kelly A. Atkins, Sarah J. Wetherill, Jelena Kusakina, Eleanor J. Walton, Maiko Kato, Miyuki Azuma, Kan Tanaka, Mitsumasa Hanaoka and Antony N. Dodd (2013) Circadian Control of Chloroplast Transcription by a Nuclear-Encoded Timing Signal. Science 339, 1316-1319.
- 2.Mitsumasa Hanaoka, Naoki Takai, Norimune Hosokawa, Masayuki Fujiwara, Yuki Akimoto, Nami Kobori, Hideo Iwasaki, Takao Kondo and Kan Tanaka (2012) RpaB, another response regulator operating circadian clock-dependent transcriptional regulation in Synechococcus elongatus PCC 7942. J. Biol. Chem. 287, 26321-26327.
- 3.Yuki Kobayashi, Sousuke Imamura, Mitsumasa Hanaoka and Kan Tanaka (2011) A tetrapyrrole-regulated ubiquitin ligase controls algal nuclear DNA replication. Nature Cell Biol. 13, 483-487.
- 4.Sousuke Imamura, Yu Kanesaki, Mio Ohnuma, Takayuki Inouye, Yasuhiko Sekine, Takayuki Fujiwara, Tsuneyoshi Kuroiwa and Kan Tanaka (2009) R2R3-type MYB transcription factor, CmMYB1, is a central nitrogen assimilation regulator in Cyanidioschyzon merolae. Proc. Natl. Acad. Sci. USA 106, 12548-12553.
- 5.Takashi Osanai, Masahiko Imashimizu, Asako Seki, Shusei Sato, Satoshi Tabata, Sousuke Imamura, Munehiko Asayama, Masahiko Ikeuchi and Kan Tanaka (2009) ChlH, the H subunit of Mg-chelatase, is an anti-sigma factor for SigE in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 106, 6860-6865.
- 6.Yuki Kobayashi, Yu Kanesaki, Ayumi Tanaka, Haruko Kuroiwa, Tsuneyoshi Kuroiwa and Kan Tanaka (2009) Tetrapyrrole signal as a cell cycle coordinator from organelle to nuclear DNA replication in plant cells. Proc. Natl. Acad. Sci. USA 106, 803-807.
- 7.Sousuke Imamura, Mitsumasa Hanaoka and Kan Tanaka (2008) The plant-specific TFIIB-related protein, pBrp, is a general transcription factor for RNA polymerase I. EMBO J. 27, 2317-2327.
- 8.Asako Seki, Mitsumasa Hanaoka, Yuki Akimoto, Susumu Masuda, Hideo Iwasaki and Kan Tanaka (2007) Induction of a group 2 sigma factor, RpoD3, by high light and the underlying mechanism in Synechococcus elongatus PCC 7942. J. Biol. Chem. 282, 36887-36894.
- 9.Mitsumasa Hanaoka, Kengo Kanamaru, Makoto Fujiwara, Hideo Takahashi and Kan Tanaka (2005) A Switch in RNA Polymerase Usage Mediated by tRNAGlu During Chloroplast Development. EMBO Rep. 6, 545-550.
- 10.Motomichi Matsuzaki, Osami Misumi, Tadasu Shin-I, Shinichiro Maruyama, Manabu Takahara, Shin-ya Miyagishima, Toshiyuki Mori, Keiji Nishida, Fumi Yagisawa, Keishin Nishida, Yamato Yoshida, Yoshiki Nishimura, Syunsuke Nakao, Tamaki Kobayashi, Yu Momoyama, Tetsuya Higashiyama, Ayumi Minoda, Masako Sano, Hisayo Nomoto, Kazuko Oishi, Hiroko Hayashi, Fumiko Ohta, Masako Sano, Hisayo Nomoto, Kazuko Oishi, Hiroko Hayashi, Fumiko Ohta, Satoko Nishizaka, Shinobu Haga, Sachiko Miura, Tomomi Morishita, Yukihiro Kabeya, Kimihiro Terasawa, Yutaka Suzuki, Yasuyuki Ishii, Syuichi Asakawa, Hiroyoshi Takano, Niji Ohta, Haruko Kuroiwa, Kan Tanaka, Nobuyoshi Shimizu, Sumio Sugano, Naoki Sato, Hisayoshi Nozaki, Naotake Ogasawara, Yuji Kohara and Tsuneyoshi Kuroiwa (2004) Genome sequence of the ultra-small unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653-657.
- 11.Ryohei Tanigawa, Masao Shirokane, Shin-ichi Maeda, Tatuso Omata, Kan Tanaka, and Hideo Takahashi (2002) Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc. Natl. Acad. Sci. USA 99, 4251-4255.
- 12.Kan Tanaka, Yuzuru Tozawa, Nobuyoshi Mochizuki, Kazuo Shinozaki, Akira Nagatani, Kyo Wakasa, and Hideo Takahashi (1997) Characterization of three cDNA species encoding plastid RNA polymerase sigma factors in Arabidopsis thaliana: Evidence for the sigma factor heterogeneity in higher plant plastids. FEBS Lett. 413, 309-313.
- 13.Kan Tanaka, Kosuke Oikawa, Niji Ohta, Haruko Kuroiwa, Tsuneyoshi Kuroiwa, and Hideo Takahashi (1996) Nuclear encoding of a chloroplast RNA polymerase sigma subunit in a red alga. Science 272, 1932-1935.
- 14.Kan Tanaka, Yuko Takayanagi, Nobuyuki Fujita, Akira Ishihama, and Hideo Takahashi (1993) Heterogeneity of the principal σ factor in Escherichia coli: The rpoS gene product, σ38, is a second principal σ factor of RNA polymerase in stationary-phase Escherichia coli. Proc. Natl. Acad. Sci. USA 90, 3511-3515.
- 15.Kan Tanaka, Tetsuo Shiina and Hideo Takahashi (1988) Multiple principal sigma factor homologs in eubacteria: Identification of the "rpoD Box." Science 242, 1040-1042.
Associate professor Sousuke Imamura
- 1.Imamura S. Kawase Y. Kobayashi I. Shimojima M. Ohta H. and Tanaka K. (2016) TOR (target of rapamycin) is a key regulator of triacylglycerol accumulation in microalgae. Plant Signal. Behav. 11(3) e1149285.
- 2.Imamura S. Kawase Y. Kobayashi I. Sone T. Era A. Miyagishima SY. Shimojima M. Ohta H. and Tanaka K. (2015) Target of rapamycin (TOR) plays a critical role in triacylglycerol accumulation in microalgae. Plant Mol. Biol. 89(3) 309-318.
- 3.Imamura S. Ishiwata A. Watanabe S. Yoshikawa H. and Tanaka K. (2013) Expression of budding yeast FKBP12 confers rapamycin susceptibility to the unicellular red alga Cyanidioschyzon merolae. Biochem. Biophys. Res. Commun. 439(2) 264-269.
- 4.Imamura S. Terashita M. Maruyama S. Ohnuma M. Fujita Y. Omata T. and Tanaka K. (2010) Nitrate assimilatory genes and their transcriptional regulation in a unicellular red alga Cyanidioschyzon merolae: Evidence for nitrite reduction by sulfite. Plant Cell Physiol. 51(5) 707-717.
- 5.Imamura S. Kanesaki Y. Ohnuma M. Takayuki I. Sekine Y. Fujiwara T. Kuroiwa T. and Tanaka K. (2009) R2R3-type MYB transcription factor, CmMYB1, controls the expression of nitrogen assimilation genes in response to nitrogen status in the unicellular red alga Cyanidioschyzon merolae. Proc. Natl. Acad. Sci. USA 106(30) 12548-12553.
- 6.Imamura S. Hanaoka M. and Tanaka K. (2008) The plant-specific TFIIB-related protein, pBrp, is a general transcription factor for RNA polymerase I. EMBO J. 27(17) 2317-2327.
- 7.Imamura S. Tanaka K. Shirai M. and Asayama M. (2006) Growth phase-dependent activation of nitrogen-related genes by a control network of group 1 and group 2 sigma factors in a cyanobacterium. J. Biol. Chem. 281(5) 2668-2675.
- 8.Imamura S. Asayama M. and Shirai M. (2004) In vitro transcription analysis by reconstituted cyanobacterial RNA polymerase: Roles of group 1 and 2 sigma factors and a core subunit, RpoC2. Genes Cells 9(12) 1175-1187.
- 9.Imamura S. Asayama M. Takahashi H. Tanaka K. Takahashi H. and Shirai M. (2003) Antagonistic dark/light-induced SigB/SigD, group 2 sigma factors, expression through redox potential and their roles in cyanobacteria. FEBS Lett. 554(3) 357-362.
- 10.Imamura S. Yoshihara S. Nakano S. Shiozaki N. Yamada A. Tanaka K. Takahashi H. Asayama M. and Shirai M. (2003) Purification, characterization, and gene expression of all sigma factors of RNA polymerase in a cyanobacterium. J. Mol. Biol. 325(5) 857-872.
- Research Laboratories and Subjects
- Abscisic Acid Helps Red Algae Tolerate Salt Stress by Controlling Cell Cycle Initiation | Life Science and Technology News
Contact
Professor Kan Tanaka
Room 814, R1 building, Suzukakedai campus
Email kntanaka@res.titech.ac.jp
Associate Professor Sousuke Imamura
Room 816, R1 building, Suzukakedai campus
Email simamura@res.titech.ac.jp
*Find more about the lab and the latest activities at Tanaka and Imamura Laboratory![]() .
.
*May 1, 2025:Some of the content has been updated with the latest information.