Life Science and Technology News
【Labs spotlight】Nozawa Laboratory
The Department has a variety of laboratories for Life Science and Technology, in which cutting-edge innovative research is being undertaken not only in basic science and engineering but also in the areas of medicine, pharmacy, agriculture, and multidisciplinary sciences. This "Spotlight" series features a laboratory from the Department and introduces you to the laboratory's research projects and outcomes. This time we focus on Nozawa Laboratory working on the structural analysis of 3D genome folding (Nozawa Laboratory was established in April 2022.)

Life Science and Technology
Associate Professor
Kayo Nozawa![]()
※The laboratory website was released on September 9, 2022.
| Degree | PhD 2012, The University of Tokyo |
|---|---|
| Areas of Research | Structural biology, Molecular biology, Genome biology |
| Keywords | X-ray crystallography, Cryo-electron microscopy, Genome structure, Transcriptional regulation |
| Web site | Nozawa Lab. |
Research interest
Cell type-dependent gene activation is enabled by the three-dimensional regulation of chromatin conformations, which involves "DNA-loop". DNA-loop regulates transcriptional activation with the help of a multi-subunit co-activator called Mediator (Fig. 1). Mediator directly connects activator which is bound to enhancer DNA with RNA polymerase II on the promoter DNA via a long DNA chain of several Kbps to Mbps, thereby forming the aforementioned DNA-loop structure in chromatin. A number of diseases have been linked to DNA-loop, including cancers, as DNA-loop plays a key role in forming clusters of enhancers, or super-enhancers, that drive unusually high level of expression of genes, such as oncogenes. Hence, DNA-loop may be a potential therapeutic target for selective disruption of oncogene expression. Notably, Mediator seems to have a key role in forming super-enhancer structure. Super-enhancers are clusters of DNA-loop, and they can drive high expression levels of oncogenes in cancer cells. Another interesting regulatory mechanism by Mediator is a formation of liquid-liquid phase separation (LLPS). LLPS represents a phenomenon in which proteins and/or nucleic acids achieve higher local concentrations and form “membrane-less organelles”.
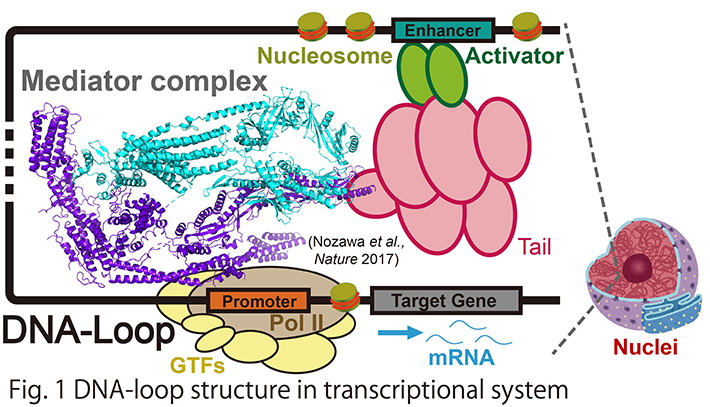
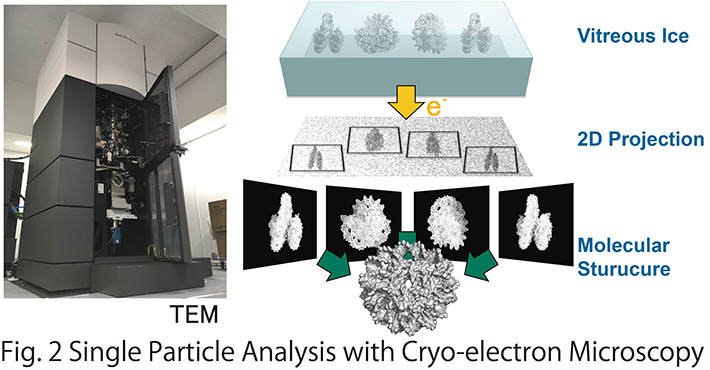
Our laboratory aims to elucidate the atomic structure of DNA-loop using Cryo-electron microscopy and X-ray crystallography, to reveal the molecular mechanism of transcriptional initiation and epigenetic regulation thereof (Fig. 2). So far, we established a protein co-expression system for the core Mediator of 15 subunits, which is the smallest functional unit of the Mediator, and performed X-ray crystallographic analysis. The result clarified that it is involved in the transcription initiation of Pol II on the promoter. We are also interested in chromatin structures constituting the DNA-loop. Applying cryo-electron microscopy analysis, we have recently characterized a unique chromatin structural unit H3-H4 octasome that resembles the normal nucleosome but lacks H2A and H2B.
Selected publications
- 1.M, Nishimura., Y, Arimura., K, Nozawa., H, Kurumizaka.
Linker DNA and histone contributions in nucleosome binding by p53
J Biochem., 168 (6), 669-675 (2020). - 2.K, Nozawa., TR, Schneider., P, Cramer.
Core Mediator structure at 3.4 Å extends model of transcription initiation complex
Nature., 545, 248–251 (2017). - 3.K, Nozawa., R, Ishitani., T, Yoshihisa., M, Sato., F, Arisaka., S, Kanamaru., N, Dohmae., D, Mangroo., B, Senger., HD, Becker., O, Nureki.
Crystal structure of Cex1p reveals the mechanism of tRNA trafficking between nucleus and cytoplasm
Nucl. Acids Res., 41 (6):3901-3914 (2013). - 4.H, Katayama., K, Nozawa., O, Nureki., S, Aimoto., Y, Nakahara., H, Hojo.
Pyrrolysine Analogs as Substrates for Bacterial Pyrrolysyl-tRNA Synthetase in Vitro and in Vivo
Bioscience, Biotechnology, and Biochemistry, 76, 110653-1-4 (2012). - 5.K, Nozawa., P, O'Donoghue., S, Gundllapalli., Y, Araiso., R, Ishitani.,
T, Umehara., D, Söll., O, Nureki.
Pyrrolysyl-tRNA synthetase-tRNA (Pyl) structure reveals the molecular basis of orthogonality
Nature., 457, 1163-1167 (2009). - 6.K, Murakami., M, Stewart., K, Nozawa., K, Tomii., N, Kudou., N, Igarashi., Y, Shirakihara., S, Wakatsuki., T, Yasunaga. & T, Wakabayashi.
Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T
Proc. Natl. Acad. Sci. U.S.A., 105, 7200-7205 (2008).
Contact
Associate Professor Kayo Nozawa
Room 707, B1B2 building, Suzukakedai campus
E-mail : nozawa.k.ab@m.titech.ac.jp
Tel / Fax : +8145-924-5727
*Find more about the lab and the latest activities at the lab site![]() .
.