Life Science and Technology News
Catching histones by the tail: a new probe to track histone modifications in living cells
Scientists at Tokyo Institute of Technology have developed a sensitive fluorescent antibody probe that specifically detects monomethylation of lysine 20 in histone H4 in living cells. This research has future implications and can be used to monitor the dynamics of histone modifications and genome integrity in single living cells without disturbing cellular functions.
Genomic integrity in living cells is maintained by packaging of nuclear DNA into chromatin, which protects it from damage and controls gene replication and expression. Histones are the primary protein components of chromatin and their post-translational modifications regulate chromatin structure and play a fundamental role in biological processes such as DNA repair, DNA replication, mitosis, etc. Among the modifications, methylation of histone H4 at lysine 20 (H4K20) is evolutionarily conserved from yeast to humans and exists in three states, mono-, di- and trimethylation, each of which have distinct biological roles. Conventional techniques studying regulation by histone modifications are limited to fixed (dead) cells, thus preventing assessment of histone modification in single, living cells.
To address this challenge, a group of scientists led by Prof. Kimura, who teaches Life Science and Technology, from the Institute of Innovative Research, Tokyo Institute of Technology, generated a genetically encoded live-cell imaging probe for sensitive monitoring of the intracellular spatiotemporal dynamics of H4K20 monomethylation (H4K20me1). The probe, called mintbody (modification-specific intracellular antibody), is a single-chain variable fragment tagged with a fluorescent protein that demonstrates high specificity for H4K20me1 over di- and trimethylation in living yeast, mammalian cells, and even multicellular organisms.
H4K20me1 is most likely associated with the tight packing of a redundant (inactivated) female X chromosome (Xi) into heterochromatin. In a roundworm Caenorhabditis elegans model, Prof. Kimura and colleagues showed that the H4K20me1-mintbody could be used to monitor changes in H4K20me1 over the cell cycle and localization of dosage-compensated X chromosomes without disrupting cell function. Thus, the new mintbody can overcome the challenges associated with visualizing and tracking histone modifications directly in living cells.
This research also identified key amino acids responsible for H4K20me1-mintbody conformational stability, solubility, and consequently, functional performance using X-ray crystallography and genetic analyses. Thus, a possible solution to the existing problem of limited solubility of intracellularly expressed antibody fragments due to aberrant folding in the cytoplasm that restricted their use was formulated.
In the future, development of additional mintbodies specific to diverse post-translational histone modifications will facilitate the identification of regulatory mechanisms that control epigenetic modifications.
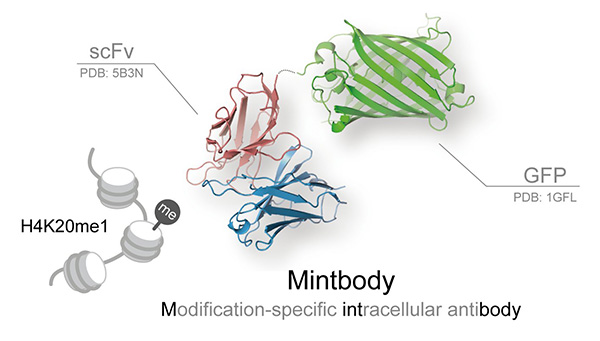
Figure. The crystal structure of H4K20me1-mintbody. Single chain variable fragment of antibody (scFv) tagged with green fluorescent protein (GFP) recognizes and binds to histone H4 post-translationally modified by the addition of a single methyl group (monomethylation) at lysine 20 (H4K20me1).
Reference
| Authors: | Yuko Sato1, Tomoya Kujirai2, Ritsuko Arai3, Haruhiko Asakawa4, Chizuru Ohtsuki4, Naoki Horikoshi2, Kazuo Yamagata5, Jun Ueda6, Takahiro Nagase7, Tokuko Haraguchi4, 8, Yasushi Hiraoka4, 8, Akatsuki Kimura3, Hitoshi Kurumizaka2 and Hiroshi Kimura1 |
|---|---|
| Title of original paper: | A Genetically Encoded Probe for Live-Cell Imaging of H4K20 Monomethylation |
| Journal: | Journal of Molecular Biology |
| DOI : | 10.1016/j.jmb.2016.08.010 |
| Affiliations : | 1 Cell Biology Unit, Institute of Innovative Research, Tokyo Institute of Technology 2 Laboratory of Structural Biology, Graduate School of Advanced Science and Engineering, Waseda University 3 Cell Architecture Laboratory, Structural Biology Center, National Institute of Genetics 4 Graduate School of Frontier Biosciences, Osaka University 5 Faculty of Biology-Oriented Science and Technology, Kindai University 6 Center for Education in Laboratory Animal Research, Chubu University 7 Public Relations Team, Kazusa DNA Research Institute 8 Advanced ICT Research Institute, National Institute of Information and Communications Technology (NICT) |
- KIMURA LAB
- Researcher Profile | Tokyo Tech STAR Search - Hiroshi Kimura
- Professor Hiroshi Kimura Receives 2015 Robert Feulgen Prize | Tokyo Tech News
- Institute of Innovative Research (IIR)
- Latest Research News
Further information
Professor Hiroshi Kimura
Cell Biology Unit, Institute of Innovative Research
Email hkimura@bio.titech.ac.jp
Tel +81-45-924-5742